Now published - link here
******
Preprint
Multimodal Molecular Imaging Reveals Tissue-Based T Cell Activation and Viral RNA Persistence for Up to Two Years Following COVID-19
Michael J Peluso; Dylan M Ryder; Robert Flavell; Yingbing Wang; Jelena Levi; Brian H LaFranchi; Tyler-Marie M Deveau; Amanda M Buck; Sadie E Munter; Kofi A Asare; Maya Aslam; Walter Koch; Gyula Szabo; Rebecca Hoh; Monika Deswal; Antonio Rodriguez; Melissa Buitrago; Viva Tai; Uttam Shrestha; Scott Lu; Sarah A Goldberg; Thomas Dalhuisen; Matthew S Durstenfeld; Priscilla Y Hsue; J D Kelly; Nitasha Kumar; Jeffrey N Martin; Aruna Gambhir; Ma Somsouk; Youngho Seo; Steven G Deeks; Zoltan G Laszik; Henry F VanBrocklin; Timothy J Henrich
The etiologic mechanisms of post-acute medical morbidities and unexplained symptoms (Long COVID) following SARS-CoV-2 infection are incompletely understood. There is growing evidence that viral persistence and immune dysregulation may play a major role. We performed whole-body positron emission tomography (PET) imaging in a cohort of 24 participants at time points ranging from 27 to 910 days following acute SARS-CoV-2 infection using a novel radiopharmaceutical agent, [18F]F-AraG, a highly selective tracer that allows for anatomical quantitation of activated T lymphocytes.
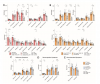
Tracer uptake in the post-acute COVID group, which included those with and without Long COVID symptoms, was significantly higher compared to pre-pandemic controls in many anatomical regions, including the brain stem, spinal cord, bone marrow, nasopharyngeal and hilar lymphoid tissue, cardiopulmonary tissues, and gut wall. Although T cell activation tended to be higher in participants imaged closer to the time of the acute illness, tracer uptake was increased in participants imaged up to 2.5 years following SARS-CoV-2 infection. We observed that T cell activation in spinal cord and gut wall was associated with the presence of Long COVID symptoms. In addition, tracer uptake in lung tissue was higher in those with persistent pulmonary symptoms. Notably, increased T cell activation in these tissues was also observed in many individuals without Long COVID.
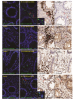
Given the high [18F]F-AraG uptake detected in the gut, we obtained colorectal tissue for in situ hybridization SARS-CoV-2 RNA and immunohistochemical studies in a subset of participants with Long COVID symptoms. We identified cellular SARS-CoV-2 RNA in rectosigmoid lamina propria tissue in all these participants, ranging from 158 to 676 days following initial COVID-19 illness, suggesting that tissue viral persistence could be associated with long-term immunological perturbations.
Link | PDF (Preprint: MedRxiv)
******
Preprint
Multimodal Molecular Imaging Reveals Tissue-Based T Cell Activation and Viral RNA Persistence for Up to Two Years Following COVID-19
Michael J Peluso; Dylan M Ryder; Robert Flavell; Yingbing Wang; Jelena Levi; Brian H LaFranchi; Tyler-Marie M Deveau; Amanda M Buck; Sadie E Munter; Kofi A Asare; Maya Aslam; Walter Koch; Gyula Szabo; Rebecca Hoh; Monika Deswal; Antonio Rodriguez; Melissa Buitrago; Viva Tai; Uttam Shrestha; Scott Lu; Sarah A Goldberg; Thomas Dalhuisen; Matthew S Durstenfeld; Priscilla Y Hsue; J D Kelly; Nitasha Kumar; Jeffrey N Martin; Aruna Gambhir; Ma Somsouk; Youngho Seo; Steven G Deeks; Zoltan G Laszik; Henry F VanBrocklin; Timothy J Henrich
The etiologic mechanisms of post-acute medical morbidities and unexplained symptoms (Long COVID) following SARS-CoV-2 infection are incompletely understood. There is growing evidence that viral persistence and immune dysregulation may play a major role. We performed whole-body positron emission tomography (PET) imaging in a cohort of 24 participants at time points ranging from 27 to 910 days following acute SARS-CoV-2 infection using a novel radiopharmaceutical agent, [18F]F-AraG, a highly selective tracer that allows for anatomical quantitation of activated T lymphocytes.
Tracer uptake in the post-acute COVID group, which included those with and without Long COVID symptoms, was significantly higher compared to pre-pandemic controls in many anatomical regions, including the brain stem, spinal cord, bone marrow, nasopharyngeal and hilar lymphoid tissue, cardiopulmonary tissues, and gut wall. Although T cell activation tended to be higher in participants imaged closer to the time of the acute illness, tracer uptake was increased in participants imaged up to 2.5 years following SARS-CoV-2 infection. We observed that T cell activation in spinal cord and gut wall was associated with the presence of Long COVID symptoms. In addition, tracer uptake in lung tissue was higher in those with persistent pulmonary symptoms. Notably, increased T cell activation in these tissues was also observed in many individuals without Long COVID.
Given the high [18F]F-AraG uptake detected in the gut, we obtained colorectal tissue for in situ hybridization SARS-CoV-2 RNA and immunohistochemical studies in a subset of participants with Long COVID symptoms. We identified cellular SARS-CoV-2 RNA in rectosigmoid lamina propria tissue in all these participants, ranging from 158 to 676 days following initial COVID-19 illness, suggesting that tissue viral persistence could be associated with long-term immunological perturbations.
Link | PDF (Preprint: MedRxiv)
Last edited by a moderator: