Circulating cell-free RNA signatures for the characterization and diagnosis of myalgic encephalomyelitis/chronic fatigue syndrome
Anne E. Gardella, Daniel Eweis-LaBolle, Conor J. Loy, Emma D. Belcher, Joan S. Lenz, Carl J. Franconi, Sally Y. Scofield, Andrew Grimson, Maureen R. Hanson, Iwijn De Vlaminck
[Line breaks added]
Significance
Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) is a debilitating illness that affects millions of individuals worldwide. Despite the growing prevalence of ME/CFS, patients suffer from the unavailability of laboratory diagnostic tests.
Here, we establish cell-free RNA (cfRNA) as a minimally invasive bioanalyte for investigating circulating biomarkers and the pathobiology of ME/CFS. Using machine learning, we develop a cfRNA-based diagnostic model with high accuracy.
We find evidence of immune system dysfunction in patients, with elevated levels of immune cell-derived transcripts as well as chronic inflammatory signaling pathways.
These findings highlight the potential of circulating cfRNA to advance biomarker discovery and uncover disease mechanisms for ME/CFS.
Abstract
People living with myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) experience heterogeneous and debilitating symptoms that lack sufficient biological explanation, compounded by the absence of accurate, noninvasive diagnostic tools.
To address these challenges, we explored circulating cell-free RNA (cfRNA) as a blood-borne bioanalyte to monitor ME/CFS. cfRNA is released into the bloodstream during cellular turnover and reflects dynamic changes in gene expression, cellular signaling, and tissue-specific processes.
We profiled cfRNA in plasma by RNA sequencing for 93 ME/CFS cases and 75 healthy sedentary controls, then applied machine learning to develop diagnostic models and advance our understanding of ME/CFS pathobiology.
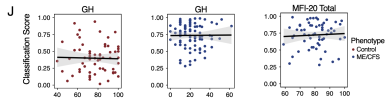
A generalized linear model with least absolute shrinkage selector operator regression trained on condition-specific signatures achieved a test-set AUC of 0.81 and an accuracy of 77%.
Immune cfRNA deconvolution revealed differences in platelet-derived cfRNA between cases and controls, as well as elevated levels of plasmacytoid dendritic, monocyte, and T cell–derived cfRNA in ME/CFS. Biological network analysis further implicated immune dysfunction in ME/CFS, with signatures of cytokine signaling and T cell exhaustion.
These findings demonstrate the utility of RNA liquid biopsy as a minimally invasive tool for unraveling the complex biology behind chronic illnesses.
Web | PNAS | Paywall
Anne E. Gardella, Daniel Eweis-LaBolle, Conor J. Loy, Emma D. Belcher, Joan S. Lenz, Carl J. Franconi, Sally Y. Scofield, Andrew Grimson, Maureen R. Hanson, Iwijn De Vlaminck
[Line breaks added]
Significance
Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) is a debilitating illness that affects millions of individuals worldwide. Despite the growing prevalence of ME/CFS, patients suffer from the unavailability of laboratory diagnostic tests.
Here, we establish cell-free RNA (cfRNA) as a minimally invasive bioanalyte for investigating circulating biomarkers and the pathobiology of ME/CFS. Using machine learning, we develop a cfRNA-based diagnostic model with high accuracy.
We find evidence of immune system dysfunction in patients, with elevated levels of immune cell-derived transcripts as well as chronic inflammatory signaling pathways.
These findings highlight the potential of circulating cfRNA to advance biomarker discovery and uncover disease mechanisms for ME/CFS.
Abstract
People living with myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) experience heterogeneous and debilitating symptoms that lack sufficient biological explanation, compounded by the absence of accurate, noninvasive diagnostic tools.
To address these challenges, we explored circulating cell-free RNA (cfRNA) as a blood-borne bioanalyte to monitor ME/CFS. cfRNA is released into the bloodstream during cellular turnover and reflects dynamic changes in gene expression, cellular signaling, and tissue-specific processes.
We profiled cfRNA in plasma by RNA sequencing for 93 ME/CFS cases and 75 healthy sedentary controls, then applied machine learning to develop diagnostic models and advance our understanding of ME/CFS pathobiology.
A generalized linear model with least absolute shrinkage selector operator regression trained on condition-specific signatures achieved a test-set AUC of 0.81 and an accuracy of 77%.
Immune cfRNA deconvolution revealed differences in platelet-derived cfRNA between cases and controls, as well as elevated levels of plasmacytoid dendritic, monocyte, and T cell–derived cfRNA in ME/CFS. Biological network analysis further implicated immune dysfunction in ME/CFS, with signatures of cytokine signaling and T cell exhaustion.
These findings demonstrate the utility of RNA liquid biopsy as a minimally invasive tool for unraveling the complex biology behind chronic illnesses.
Web | PNAS | Paywall