mariovitali
Senior Member (Voting Rights)
cc : @JaimeS ,@ScottTriGuy , @Perrier
Further to the hypothesis that ME/CFS originates from a Liver Injury caused by Viral Infection / Certain Medications / Toxic Substances here is more Research suggesting this hypothesis. The Research i present involves organophosphates and Depleted Uranium (DU). Please note that DU is thought to be the cause of Symptoms of Gulf War Ilness Syndrome researched by Nancy Klimas.
First, regarding DU :

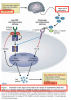
We note : Bile Acid Metabolism disruption, involvement of CYP27A1 metabolites and LXR which can be seen in the Network Analysis graph (apart from FXR, LXR) created in 2017 and was also part of my presentation in EUROMENE :

Now, Organophosphates :

There was a Thread on Phoenix Rising regarding Organophosphates mentioning a plane crash in Holland. Among its cargo where organophosphates and depleted uranium was contained in the tail of the aircraft.
From Wikipedia :
and
Links :
https://forums.phoenixrising.me/ind...e-dichlorvos-and-mitochondria-toxicity.54754/
https://en.wikipedia.org/wiki/El_Al_Flight_1862#Cargo
Further to the hypothesis that ME/CFS originates from a Liver Injury caused by Viral Infection / Certain Medications / Toxic Substances here is more Research suggesting this hypothesis. The Research i present involves organophosphates and Depleted Uranium (DU). Please note that DU is thought to be the cause of Symptoms of Gulf War Ilness Syndrome researched by Nancy Klimas.
First, regarding DU :

We note : Bile Acid Metabolism disruption, involvement of CYP27A1 metabolites and LXR which can be seen in the Network Analysis graph (apart from FXR, LXR) created in 2017 and was also part of my presentation in EUROMENE :

Now, Organophosphates :
There was a Thread on Phoenix Rising regarding Organophosphates mentioning a plane crash in Holland. Among its cargo where organophosphates and depleted uranium was contained in the tail of the aircraft.
From Wikipedia :
After about a year, however, many residents and service personnel began approaching doctors with physical health complaints, which the affected patients blamed on the El Al crash. Insomnia, chronic respiratory infections, general pain and discomfort, impotence, flatulence, and bowel complaints were all reported. 67% of the affected patients were found to be infected with Mycoplasma, and suffered from symptoms similar to the Gulf War Syndrome or Chronic Fatigue Syndrome-like symptoms.
and
The first studies on the symptoms reported by survivors, performed by the Academisch Medisch Centrum, began in May 1998. The AMC eventually concluded that up to a dozen cases of auto-immune disorders among the survivors could be directly attributed to the crash, and health notices were distributed to doctors throughout the Netherlands requesting that extra attention be paid to symptoms of auto-immune disorder, particularly if the patient had a link with the Bijlmer crash site.
Links :
https://forums.phoenixrising.me/ind...e-dichlorvos-and-mitochondria-toxicity.54754/
https://en.wikipedia.org/wiki/El_Al_Flight_1862#Cargo