On the topic of the brain expression, I don't remember much discussion about this yet. While all 13 brain tissues had enrichment of ME/CFS genes, there is an ordering of most to least significant that might give some clues.
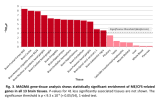
Written out and grouped:
So genes associated with ME/CFS tend to be the genes relatively more expressed than other genes in the brain. And this seems to be most prominent in the cortex/frontal cortex and least prominent in the pituitary gland. Does this ordering mean anything? Are the ones near the top maybe more associated with "higher order" functions?

Written out and grouped:
I added pituitary gland even though it didn't reach the significance threshold since it's the one other brain-related tissue they tested against.
- High
- Brain Frontal Cortex BA9: ~8.3
- Brain Cortex: ~8.0
- Brain Anterior cingulate cortex BA24: ~7.9
- Medium
- Brain Nucleus accumbens basal ganglia: ~7.0
- Brain Caudate basal ganglia: ~6.2
- Brain Amygdala: ~6.1
- Brain Hippocampus: ~6.0
- Brain Cerebellar Hemisphere: ~6.0
- Brain Hypothalamus: ~5.9
- Brain Cerebellum: ~5.8
- Brain Putamen basal ganglia: ~5.5
- Low
- Brain Spinal cord cervical c-1: ~3.9
- Brain Substantia nigra: ~3.6
- Pituitary: ~2.5
So genes associated with ME/CFS tend to be the genes relatively more expressed than other genes in the brain. And this seems to be most prominent in the cortex/frontal cortex and least prominent in the pituitary gland. Does this ordering mean anything? Are the ones near the top maybe more associated with "higher order" functions?
Last edited:
