Many thanks. Try as I might, I cannot find anything about non-coding genes. The paper states "There were 43 protein-coding genes with at least one eQTL within an ME/CFS genome-wide significant interval, and we prioritised 29 ME/CFS candidate causal genes among them..." which sounds like they only looked at protein-coding genes.I'm not sure that is the case. The first thing they look for is the genetic signal, then they look to see what was captured by that genetic signal. I think that was mainly protein coding genes. I had a feeling there was at least one RNA species. I don't know if that showed up in the supplementary information?
You are using an out of date browser. It may not display this or other websites correctly.
You should upgrade or use an alternative browser.
You should upgrade or use an alternative browser.
Preprint Initial findings from the DecodeME genome-wide association study of myalgic encephalomyelitis/chronic fatigue syndrome, 2025, DecodeMe Collaboration
wigglethemouse posted them a few posts back. These are in order of significance, with most significant at the top.Also, MAG highlights 13 genes. Not sure if the supplementary information lists the 13? That would be interesting to see.
LRRC7
STAU1
CSE1L
DARS2
ZBTB37
TAOK3
ARFGEF2
DNAH10OS
ZNF664
CCDC92
HIST1H4H
ZNF311
SUDS3
Yann04
Senior Member (Voting Rights)
Is the 5k Lipkin GWAS powered strong enough to have a decent chance at replicating Decode? From what I understand if it’s done genome wide with multiple test corrections 5k is unlikely to meet significance thresholds.
I assume there’s other values, Like hypothesis driven analysis so you don’t have to multiple test correct the rest of the SNPs, and other statistics that can be used to evaluate the consistency of findings with Decode.
I assume there’s other values, Like hypothesis driven analysis so you don’t have to multiple test correct the rest of the SNPs, and other statistics that can be used to evaluate the consistency of findings with Decode.
Andy
Senior Member (Voting rights)
Meta analysis, especially if it has the same selection criteria as DecodeME.Is the 5k Lipkin GWAS powered strong enough to have a decent chance at replicating Decode? From what I understand if it’s done genome wide with multiple test corrections 5k is unlikely to meet significance thresholds.
I assume there’s other values, Like hypothesis driven analysis so you don’t have to multiple test correct the rest of the SNPs, and other statistics that can be used to evaluate the consistency of findings with Decode.
@wigglethemouse has taken up the MAGMA baton, thankfully. Meanwhile this non-scientist is trying to make sense of the MAGMA paper - if you experts can explain what is going on here, it will avoid me repeatedly using my forehead...Why is the MAGMA analysis of seemingly little interest in this thread? Do people not like MAGMA analysis? The results (all brain tissues) seemed very interesting to my non-scientist eyes.
Utsikt
Senior Member (Voting Rights)
The search function allows you to search just the thread you’re in. Here are the results for «sleep» in this thread:24 pages in a couple days of being afkcannot conceivably go through all this, please excuse my question:
Have any findings been linked to sleep disruption?
Search results for query: Sleep
I don't think that's what's happening though. I'm pretty sure the following is right, but it's hard to find good explanations.The MAGMA analysis seems to take 13 genes from the SNP comparison but this seems like an informed guess. We don't know if those 13 genes are really implicated. Many don't match with the FUMA/coloc analysis.
MAGMA is fully separate from the part where they selected candidate genes based on things like eQTLs and nearness. Instead they take every gene and assign it a score based on how significantly the SNPs within and around it were associated with ME/CFS in the GWAS. At this point it outputs that 13 genes are significantly associated with ME/CFS. I agree at this point, you still don't know which, if any, of these 13 genes are actually causal for ME/CFS. They're just significantly associated, but there's still multiple per locus so some are probably just correlated to a real gene.
But the tissue part doesn't use just the 13 genes. It takes the score for every single gene and compares it to the expression level in a tissue. A simple way to think about it: You take one tissue, say the pituitary gland. You make a plot that includes every one of the ~18,000 genes. On the x-axis is the MAGMA score, in other words how associated were the SNPs around the gene with ME/CFS. On the y-axis is the expression level of that gene in the pituitary gland. If there's a positive slope, you say ME/CFS genes are enriched in the pituitary gland. And you look at this slope for every tissue. I think the actual method includes covariates, but that's the gist.
So while some or all of the 13 top genes might potentially be wrong, the tissue analysis includes every gene. There's an assumption that throughout the DNA there will be hundreds or thousands of genes that only have a tiny barely detectable effect on ME/CFS, but if you include them all and look at what tissues all these hundreds or thousands of tiny and large effect genes are enriched in, a signal might emerge. And here it emerged in the brain. If the scores were random, you wouldn't expect a positive slope in any tissue.
Some documents I looked at to try to understand it:
This tells MAGMA to perform annotation, mapping SNPs to genes based on the transcription region of each gene. A SNP is mapped to a gene if it is located either inside the transcription region of the gene, or in a window around it. In this case we specify the window to reach up to 1 kilobase upstream of the transcription start site, and 0.5 kilobases downstream of the transcription stop site.
Dynamic expression of genes associated with schizophrenia and bipolar disorder across development - PMC
Common genetic variation contributes a substantial proportion of risk for both schizophrenia and bipolar disorder. Furthermore, there is evidence of significant, but not complete, overlap in genetic risk between the two disorders. It has been ...
Briefly, common SNP association P-values were combined into gene-wide P-values (via the MAGMA SNP-wise mean model), using a window of 35 kb upstream and 10 kb downstream43 of each gene in order to include SNPs within regulatory regions. Only protein-coding genes were included in the analysis.
The gene property analysis method performs a linear regression of gene-wide association against a gene-level property (here, relative expression score), in which covariates are included to correct for potential confounds.
Is this what they did:@Chris Ponting Would you be kind enough to help us interpret the MAGMA analysis paragraph in the paper (discussion in above post).
Are the 13 genes in table S4 found from gene based analysis only and then tested for tissue, or is the list a gene-tissue enrichment analysis presented as a gene set - table S4 and then tested against all tissue types in Fig 3. Or is fig 3 showing something different? Which analysis is the last sentence on significance referring too - is it a tissue one or non-tissue one?
a) identify gene-sets for each of 54 tissues (from GO? where?)
b) MAGMA test: for each such gene-set, do member genes (out of the total 18,637 genes) have, on average, lower p-values than all other non-gene-set genes
c) only 13 genes belonged to these MAGMA gene-sets
I thought it'd be interesting to compare the DecodeME result for the tissue analysis to other papers. I searched for "MAGMA tissue" in Google Images, DuckDuckGo Images, and searched in Google Scholar to find other papers that included plots like the one here.
Panic disorder:
Brain regions are near the top, but none were significant after correction.
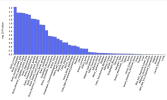
PTSD:
Also here, brain regions are all significanly enriched. Even including pituitary in this case.
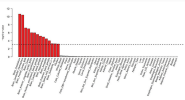
Loneliness:
Brain again.

Varicose veins:
Nice, as one would expect, brain is not significant here, but arteries are near the top. Plus things like uterus and breast tissue.
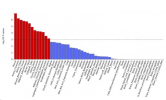
Blood pressue:
Interestingly, brain.

Ulcerative colitis:
Some of the most significant hits were spleen, bladder, small intestine, and lung.
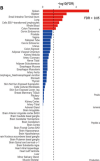
To not have too many images, I'll just write out what the plot shows for the rest:
Circadian rhythm (sleep timing): Brain
Skeletal muscle mass: Brain
Sleep duration: Brain
Anxiety: Brain
Falling risk: Brain
Non-alcoholic fatty liver disease: Liver
Coronary artery disease: Arteries, uterus, esophagus, etc
Cystatin-C kidney function: Kidney, pancreas, stomach, etc
So while there are a couple sort of surprising ones like skeletal muscle mass genes being encriched in the brain, they mostly seem to make sense.
Panic disorder:
Brain regions are near the top, but none were significant after correction.

PTSD:
Also here, brain regions are all significanly enriched. Even including pituitary in this case.

Loneliness:
Brain again.

Varicose veins:
Nice, as one would expect, brain is not significant here, but arteries are near the top. Plus things like uterus and breast tissue.

Blood pressue:
Interestingly, brain.

Ulcerative colitis:
Some of the most significant hits were spleen, bladder, small intestine, and lung.

To not have too many images, I'll just write out what the plot shows for the rest:
Circadian rhythm (sleep timing): Brain
Skeletal muscle mass: Brain
Sleep duration: Brain
Anxiety: Brain
Falling risk: Brain
Non-alcoholic fatty liver disease: Liver
Coronary artery disease: Arteries, uterus, esophagus, etc
Cystatin-C kidney function: Kidney, pancreas, stomach, etc
So while there are a couple sort of surprising ones like skeletal muscle mass genes being encriched in the brain, they mostly seem to make sense.
You are (way) ahead of me - I was reading the original 2015 MAGMA paper where the gene-set variables are just 0/1 although those authors point out "The variables C1, C2, . . ., in this generalized gene-set analysis model can reflect any gene property, from the binary indicators used for the competitive gene-set analysis to continuous variables such as gene size and expression levels."It takes the score for every single gene and compares it to the expression level in a tissue.
Discussion is underway on webinar. Livestreaming on Facebook
 www.facebook.com
www.facebook.com
Log in to Facebook
Log in to Facebook to start sharing and connecting with your friends, family and people you know.
 www.facebook.com
www.facebook.com
ME/CFS Science Blog
Senior Member (Voting Rights)
Thanks, will try to take a closer look at this. So it's like they used all possible genes but weighed them by how much the SNP signal from the GWAS points to them? That would make more sense and make there results more interesting.So while some or all of the 13 top genes might potentially be wrong, the tissue analysis includes every gene.
Yes, that's how I understand it. For each gene, they look at all the many SNPs that are in or around that area of the DNA where the gene's code is located to give that gene a score based on how significant those nearby SNPs were in the GWAS (how high they are in the manhattan plot). They account for linkage disequilibrium between SNPs to not "double-count" a genetic signal in a gene's score if multiple SNPs are all significant together just because of LD.So it's like they used all possible genes but weighed them by how much the SNP signal from the GWAS points to them? That would make more sense and make there results more interesting.
ME/CFS Science Blog
Senior Member (Voting Rights)
I think in the DecodeME webinar just now Sonya said that they had 9 requests from other researchers to access the DecodeME data.
ME/CFS Science Blog
Senior Member (Voting Rights)
In that case it might be quite important. I wonder if we should interpret the likelihood of possible genes in light of this MAGMA analysis: those that are not expressed in the brain might be less likely to be a relevant gene compared to those who are highly expressed in the brain (Figure 4 In the paper)?For each gene, they look at all the many SNPs that are in or around that area of the DNA where the gene's code is located to give that gene a score based on how significant those nearby SNPs were in the GWAS (how high they are in the manhattan plot)
Hmm. I think on its face that does make sense (though not sure how much confidence this actually allows us to have in selecting genes based on expression). I wonder if any of the papers I linked above that did MAGMA tissue analyses did anything similar. I didn't read any yet, just grabbed the plots.In that case it might be quite important. I wonder if we should interpret the likelihood of possible genes in light of this MAGMA analysis: those that are not expressed in the brain might be less likely to be a relevant gene compared to those who are highly expressed in the brain (Figure 4 In the paper)?
Edit: I'm just thinking it still might be possible that one of the significant loci has to do with another part of the body, so I'm worried about dismissing a non brain gene prematurely.
I'm thinking maybe the plot shows that the brain is important, but doesn't necessarily conclusively say which parts of the body are not important.
Testes, EBV-transformed lymphocytes, and muscle are next highest after brain (though not significant after adjustment). The second two make some sense as well for ME/CFS.
Edit 2: Actually looks like they're not even significant before correction. Both around p of 0.1).
Last edited:
jnmaciuch
Senior Member (Voting Rights)
my feeling is that it's really early for anyone to be saying with much confidence that the genes they found point to any specific pathway.
My feeling is that it’s worth making a distinction between “these identified genes have previously been found to be important in immune and nervous system function, suggestion that those broad areas are interesting directions for future focus” and “these results show that the illness is driven by neuro-immune pathways.”I tend to agree. For most of the loci there are multiple potential genes implicated and each of the genes are involved in multiple pathways.
You’re both definitely right that those genes are not exclusively expressed in either system, and we have no way of knowing which functions of those genes are relevant. Or if they are relevant in maintaining disease state or indirectly predisposing to its trigger.
And like you said @forestglip , anyone with a bit of time on their hands could probably link several of these genes to spin whatever story they wanted. Hell, AI can do it for you.
I do also agree with @chillier that there’s a need to generate good testable hypotheses. In which case these genes would be an additional piece of evidence in favor of a hypothesis, but the hypothesis should also be able to stand on its own.
I certainly don’t think these results should be used to exclude a viable hypothesis if other evidence or reasoning points in its favor—trying to push all ME/CFS research into a strictly neuro-immune boat solely on the basis of these results would be shooting ourselves in the foot. But it does make that look like a more promising direction overall.
wigglethemouse
Senior Member (Voting Rights)
I think the GWAS arrays were specifically designed to focus on protein coding genes. Just like Whole Exome Sequencing. It's only relatively recently that Whole Genome Sequencing has become more competitively priced for full coverage, but still more expensive than GWAS arrays.Many thanks. Try as I might, I cannot find anything about non-coding genes. The paper states "There were 43 protein-coding genes with at least one eQTL within an ME/CFS genome-wide significant interval, and we prioritised 29 ME/CFS candidate causal genes among them..." which sounds like they only looked at protein-coding genes.
How are we to reconcile the somewhat different genes and tissues of:
Fig. 3. MAGMA gene-tissue analysis shows statistically significant enrichment of ME/CFS-related genes in all 13 brain tissues.
Fig. 4: Approximate Bayes factor posterior probability (PPH4) that mRNA expression and ME/CFS traits are associated and share a single causal variant.
Fig. 3. MAGMA gene-tissue analysis shows statistically significant enrichment of ME/CFS-related genes in all 13 brain tissues.
Fig. 4: Approximate Bayes factor posterior probability (PPH4) that mRNA expression and ME/CFS traits are associated and share a single causal variant.
I think the GWAS arrays were specifically designed to focus on protein coding genes.
Thanks so much. Presumably the vast non-coding genome regions would contain very few of the study's GWAS genotyped markers, so that the "Phasing and genotype imputation" genotyping steps would not not impute anything statistically reliable.
