hotblack
Senior Member (Voting Rights)
That’s very positive news! Thanks for doing this @Jonathan Edwards
And good probing questions @voner
And good probing questions @voner
what did you learn that can be shared with us?
Which is really good news!That some other people think this is really interesting!
Or is it more of a pointer to the general pathology
SourceEnhancers and promoters are gene-regulatory elements. They are stretches of DNA that help in both eukaryotic and prokaryotic transcription. The promoters are known to initiate transcription, and the enhancers increase the level of transcription
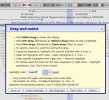
Thanks! The highlighting doesn’t seem very touchscreen friendly unfortunately. But the rest seems to work, I’ll have an explore.@hotblack you might like to explore some of the tracks on UCSC genome browser--it compiles a lot of this information visually so you can do some exploring.
For instance pick the top one, a promoter/enhancer with a high score (it has a little star by it to show this too)
Expand the info and see the location of it is chr17:52158438-52159092
You can zoom in to that location on the DecodeME LocusZoom data and see lots of the hits tie in with this area
Thanks for the tip!If you start zoomed in on a SNP, you can also hit the "highlight" button down here which will highlight everything in your viewer
Good to know I’m not talking nonsense! And nice to start to understand it a bit more.This is more or less exactly what my genetics friends at UCL did when they gave me a presentation of why they thought it was worth picking CA10 for a basic biology project.
Ensembl pipelines are very complex and it would be impossible to provide explanations for all changes between the releases. Sometimes it's possible to figure out or speculate the reasons by looking into intermediate outputs that people who ran the pipelines have but even that is quite challenging. The sheer amount of data and analyses that go into each release... It takes 3-4 or more months and something like 80-100 people working full-time to get a release out. The documentation on the website could be better, though, but it still wouldn't be enough to determine the exact reason behind each change.Wasn't kept in GRCh38, yes--unfortunately Ensembl doesn't really have detailed annotation for why certain genes get dropped in the newest release. Sometimes it's because the gene mapping is suspect, sometimes it's for some other logistical reason. I think that happens a lot to snoRNAs and miRNAs in particular just because of the sheer number of them. But that was the reasoning for creating the Archive--the current version is curated with the best intentions, but shouldn't be considered the end all be all.
The variants are in the current Ensembl release: rs11079993, rs967823. When I clicked on "Phenotype data", an additional piece of info (after a table of associations) said "This variant has not been mapped to any Ensembl genes." for both of them.[
{
"Loci ": "CA10; snoZ178",
"Publication Loci ": "CA10; snoZ178",
"Variants ": "rs11079993",
"allele 1 ": "G",
"allele 2 ": "T",
"direction ": "down",
"Phenotype ": "Other Clinical Pain",
"PMID ": "PMID:33830993",
"comments ": "Significantly associated with multisite chronic pain in female"
},
{
"Loci ": "snoZ178",
"Publication Loci ": "snoZ178",
"Variants ": "rs967823",
"allele 1 ": "G",
"allele 2 ": "A",
"direction ": "no direction reported",
"Phenotype ": "Pain",
"PMID ": "PMID:37844115",
"comments ": "Significntly ssocited with pain"
}
]
