DecodeME candidate ME gene
From the Candidate Genes document
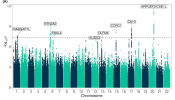
Chr17 contained one Tier 1 gene.
****************
CA10 (Tier 1)
• Protein: Carbonic anhydrase-related protein 10. UniProt. GeneCards.
Note that this gene encodes a protein that lacks catalytic activity. The allele that increases the risk of ME/CFS is associated with increased CA10 expression in prostate.
• Molecular function: CA10 binds both α- and β-neurexins, a major class of presynaptic cell adhesion molecules, and enhances their surface transport (62). CA10 is most highly expressed in the cerebellum, followed by the spinal cord and cerebral cortex, and is localised to synapses (62). CA10 inhibits the addition to neurexins of heparan sulfate (HS) glycosaminoglycan, a post-translation modification that controls the binding of presynaptic neurexins to postsynaptic neuroligins (63,64). This trans-synaptic interaction is crucial for synaptic transmission and plasticity.
In a rat model of nerve injury (spinal nerve ligation [SNL]-associated allodynia in the ipsilateral hind paw), inhibition of such Nrx1b-Neuroligin-1 interactions prevents pain (65). Nrx1β/Nrx1b contributes to neuropathic injury–induced nociceptive hypersensitivity by interacting with neurologin-1 (NL1), which subsequently activates the spinal NL1/PSD-95/pNR2B cascade. Neurexin 2 appears to be involved in pain processing (66,67).
• Cellular function: CA10 is thought to play a role in the central nervous system, especially in brain development.
• Link to disease: Tissue injury-induced synaptic redistribution of NLGN1 appears to be involvedin the development of pain hypersensitivity (68). NLGN1 in the spinal cord dorsal horn (a key site for processing pain signals) is involved in the processing of nociceptive signals. Peripheral inflammation rapidly enhances the synaptic localisation of NLGN1. Peripheral inflammation induces synaptic accumulation of the NMDA receptor. NLGN1 knockdown attenuates this accumulation and alleviates pain hypersensitivity (68).
• Potential relevance to ME/CFS: Altered CA10 expression modulates addition of heparan sulfate to presynaptic neurexins. This changes their affinity for post-synaptic neurologins, cell-cell signalling across the synapse and individuals’ pain sensitivity.
****************
References
From the Candidate Genes document
CHROMOSOME 17In DecodeME, we attempted to link GWAS variants to target genes. Here we discuss the top two tiers of predicted linked genes that we are most confident about –‘Tier 1’ and ’Tier 2’.
We defined genes as Tier 1 genes if: (i) they are protein-coding genes, (ii) they have GTEx-v10 expression quantitative trait loci (eQTLs) lying within one of the FUMA-defined ME/CFS-associated intervals, and (iii) their expression and ME/CFS risk are predicted to share a single causal variant with a posterior probability for colocalisation (H4) of at least 75%. For this definition, we disregarded the histone genes in the chr6p22.2 HIST1 cluster, as their sequences and functions are highly redundant (1). This prioritisation step yielded 29 Tier 1 genes.
For the intervals without Tier 1 genes, three Tier 2 genes were defined as the closest protein-coding genes without eQTL association: FBXL4 (chr6q16.1), OLFM4 (chr13q14.3), and CCPG1 (chr15q21.3).
Chr17 contained one Tier 1 gene.
****************
CA10 (Tier 1)
• Protein: Carbonic anhydrase-related protein 10. UniProt. GeneCards.
Note that this gene encodes a protein that lacks catalytic activity. The allele that increases the risk of ME/CFS is associated with increased CA10 expression in prostate.
• Molecular function: CA10 binds both α- and β-neurexins, a major class of presynaptic cell adhesion molecules, and enhances their surface transport (62). CA10 is most highly expressed in the cerebellum, followed by the spinal cord and cerebral cortex, and is localised to synapses (62). CA10 inhibits the addition to neurexins of heparan sulfate (HS) glycosaminoglycan, a post-translation modification that controls the binding of presynaptic neurexins to postsynaptic neuroligins (63,64). This trans-synaptic interaction is crucial for synaptic transmission and plasticity.
In a rat model of nerve injury (spinal nerve ligation [SNL]-associated allodynia in the ipsilateral hind paw), inhibition of such Nrx1b-Neuroligin-1 interactions prevents pain (65). Nrx1β/Nrx1b contributes to neuropathic injury–induced nociceptive hypersensitivity by interacting with neurologin-1 (NL1), which subsequently activates the spinal NL1/PSD-95/pNR2B cascade. Neurexin 2 appears to be involved in pain processing (66,67).
• Cellular function: CA10 is thought to play a role in the central nervous system, especially in brain development.
• Link to disease: Tissue injury-induced synaptic redistribution of NLGN1 appears to be involvedin the development of pain hypersensitivity (68). NLGN1 in the spinal cord dorsal horn (a key site for processing pain signals) is involved in the processing of nociceptive signals. Peripheral inflammation rapidly enhances the synaptic localisation of NLGN1. Peripheral inflammation induces synaptic accumulation of the NMDA receptor. NLGN1 knockdown attenuates this accumulation and alleviates pain hypersensitivity (68).
• Potential relevance to ME/CFS: Altered CA10 expression modulates addition of heparan sulfate to presynaptic neurexins. This changes their affinity for post-synaptic neurologins, cell-cell signalling across the synapse and individuals’ pain sensitivity.
****************
References
62 Sterky FH, Trotter JH, Lee SJ, Recktenwald C V, Du X, Zhou B, et al. Carbonic anhydrase-related protein CA10 is an evolutionarily conserved pan-neurexin ligand. Proc Natl Acad Sci US A. 2017 Feb 14;114(7):E1253–62.
63 Montoliu-Gaya L, Tietze D, Kaminski D, Mirgorodskaya E, Tietze AA, Sterky FH. CA10 regulates neurexin heparan sulfate addition via a direct binding in the secretory pathway. EMBO Rep. 2021 Apr 7;22(4):e51349.
64 Zhang P, Lu H, Peixoto RT, Pines MK, Ge Y, Oku S, et al. Heparan Sulfate Organizes Neuronal Synapses through Neurexin Partnerships. Cell. 2018 Sep 6;174(6):1450-1464.e23.
65 Lin TB, Lai CY, Hsieh MC, Jiang JL, Cheng JK, Chau YP, et al. Neuropathic Allodynia Involves Spinal Neurexin-1β-dependent Neuroligin-1/Postsynaptic Density-95/NR2B Cascade in Rats.Anesthesiology. 2015 Oct;123(4):909–26.
66 Xu L, Feng Q, Deng H, Zhang X, Ni H, Yao M. Neurexin-2 is a potential regulator of inflammatory pain in the spinal dorsal horn of rats. J Cell Mol Med. 2020 Dec;24(23):13623–33.
67 Xu L, Zheng S, Chen L, Yang L, Zhang S, Liu B, et al. N4-acetylcytidine acetylation of neurexin 2 in the spinal dorsal horn regulates hypersensitivity in a rat model of cancer-induced bone pain. Mol Ther Nucleic Acids. 2024 Jun 11;35(2):102200.
68 Zhao JY, Duan XL, Yang L, Liu JP, He YT, Guo Z, et al. Activity-dependent Synaptic Recruitment of Neuroligin 1 in Spinal Dorsal Horn Contributed to Inflammatory Pain. Neuroscience. 2018Sep 15;388:1–10.
69 Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997 Sep 19;90(6):1061–71.