Hi,
I developed the
MetaME and
DecodeME repositories on GitHub. I am not a scientist, just a patient.
MetaME is a meta-analysis of approximately 21,500 ME cases (DecodeME + UK Biobank + Million Veteran Program). It uses standard methodologies (METAL + FUMA). No fine-mapping was performed.
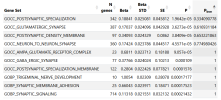
The analysis identifies significant associations between the genetic profile of this cohort and:
- a Gene Ontology term related to glutamatergic synapses,
- gene expression in several brain regions, and
- gene expression in glutamatergic neurons, both in mice and in humans.
I am still working on the DecodeME fine-mapping. As shown in the repository, I was able to use LD matrices from the UK Biobank. However, the fine-mapping currently available in the repository only employed approximately 5 million SNPs from the original DecodeME summary statistics.
I am intrigued by the possibility that ME/CFS may be linked to a dysfunction of glutamatergic signaling in the brain, at least in a subgroup. In particular, I wonder whether a deficit in this system could explain fatigue and cognitive impairment.
There is a drug used in epilepsy that antagonizes the AMPA receptor: perampanel. It is known to cause fatigue, but it does not appear to be associated with exercise intolerance. If the glutamatergic hypothesis were correct, one might expect this drug to induce an ME/CFS-like phenotype.
I would like to add more patients to my meta analysis, unfortunately I used all the available summary statistics, to my knowledge. I recently added 6,000 Long Covid patients and I found two risk loci: the one on chromosome 20, already present in DecodeME, and a new one on chromosome 22. But I am still going through this. On the other hand, when adding LC, the signal from the glutamatergic synapses, while still present, does not reach statistical significance, after Bonferroni correction (see table).