ME/CFS Science Blog
Senior Member (Voting Rights)
The DecodeME questionnaire asked about 'Clinical depression' as one of the other conditions participants might have. It would be interesting to see if the answer to this question determines the similarity to the depression GWAS. In other words, if we see similar results in ME/CFS patients, if those with clinical depression are excluded.
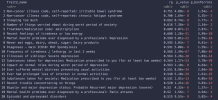
Here's an example of what they found in the first big depression GWAS. Mostly genes involved in neurons and synapses, some pointing to regulation of immune responses and also calcium channel activity.

Source: Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression | Nature Genetics
Here's an example of what they found in the first big depression GWAS. Mostly genes involved in neurons and synapses, some pointing to regulation of immune responses and also calcium channel activity.

Source: Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression | Nature Genetics