Some discussion about variants near SUDS3/TAOK3 and their possible relationship to lupus have been moved to: Genetics: Chromosome 12: SUDS3, TAOK3
You are using an out of date browser. It may not display this or other websites correctly.
You should upgrade or use an alternative browser.
You should upgrade or use an alternative browser.
Preprint Initial findings from the DecodeME genome-wide association study of myalgic encephalomyelitis/chronic fatigue syndrome, 2025, DecodeMe Collaboration
mariovitali
Senior Member (Voting Rights)
@forestglip I am not sure if you have seen this analysis by Paolo Maccallini :
https://github.com/paolomaccallini-hub/MetaME?tab=readme-ov-file
I am particularly interested in genes EP300 and UGP2. Can these two be significant targets?
https://github.com/paolomaccallini-hub/MetaME?tab=readme-ov-file
I am particularly interested in genes EP300 and UGP2. Can these two be significant targets?
Interesting that in this meta-analysis, there actually is a significant gene set enrichment in MAGMA: GOCC_GLUTAMATERGIC_SYNAPSE@forestglip I am not sure if you have seen this analysis by Paolo Maccallini :
https://github.com/paolomaccallini-hub/MetaME?tab=readme-ov-file
I am particularly interested in genes EP300 and UGP2. Can these two be significant targets?
Paolo commented this on ME/CFS Science Blog's blog about overlapping controls, so I'm not sure how much impact this may have had on the results:
There is a problem though: DecodeME and UK Biobank have overlapping controls. Now I am trying to solve this problem using a correction of the weights used in the meta analysis.
I don't know enough about the method used for the meta-analysis calculation to really comment on it. But assuming it's valid, there are still 27 resulting candidate genes at the chr22 locus which includes EP300. So that could be a gene of interest, or maybe it's another of those many gene options. And there are four candidate genes at the chr2 locus that includes UGP2.
mariovitali
Senior Member (Voting Rights)
Interesting that in this meta-analysis, there actually is a significant gene set enrichment in MAGMA: GOCC_GLUTAMATERGIC_SYNAPSE
Paolo commented this on ME/CFS Science Blog's blog about overlapping controls, so I'm not sure how much impact this may have had on the results:
I don't know enough about the method used for the meta-analysis calculation to really comment on it. But assuming it's valid, there are still 27 resulting candidate genes at the chr22 locus which includes EP300. So that could be a gene of interest, or maybe it's another of those many gene options. And there are four candidate genes at the chr2 locus that includes UGP2.
Thanks @forestglip , yes I wonder if a key mechanism here is excitotoxicity via glutamate (and perhaps quinolinic acid). Another interesting finding can potentially be the gene ACO2 (exists also in the list of genes) which could be linked to the itaconate shunt hypothesis. If we have indeed issues with glutamatergic synapses coupled with ER stress and impaired ER autophagy (=ER-phagy) in the Endoplasmic reticulum then we may have a perfect storm taking place.
Here's an attempt to bring together all of the main candidate genes from different sources: Tier 1 genes, tier 2 genes, genes significant in MAGMA, and the gene (or two genes if it is not clear) closest to a locus for the top 25 loci.
Sources:
Tier 1 and 2 genes: Candidate genes document
MAGMA genes: Supplementary table 4
Top 25 loci: Supplementary table 3, with nearest gene to each lead variant found with LocusZoom. (Can also be done on UCSC Genome Browser.)
Gene Method CHR Lead variant position (GRCh38) Reference allele Effect allele Lead variant ID Lead variant p-value LRRC7 MAGMA 1 69696474 A G 1:69696474:A:G 2.06E-07 xxxxx NEGR1 Nearest (1 of 2) 1 73126414 C CA 1:73126414:C:CA 1.19E-07 LRRIQ3 Nearest (1 of 2) 1 73126414 C CA 1:73126414:C:CA 1.19E-07 xxxxx ZNF644 Nearest 1 91028158 C T 1:91028158:C:T 1.89E-07 xxxxx RABGAP1L Tier 1 1 173846152 T C 1:173846152:T:C 2.56E-08 DARS2 Tier 1, MAGMA, Nearest 1 173846152 T C 1:173846152:T:C 2.56E-08 RC3H1 Tier 1 1 173846152 T C 1:173846152:T:C 2.56E-08 GPR52 Tier 1 1 173846152 T C 1:173846152:T:C 2.56E-08 ZBTB37 Tier 1, MAGMA 1 173846152 T C 1:173846152:T:C 2.56E-08 TNFSF4 Tier 1 1 173846152 T C 1:173846152:T:C 2.56E-08 ANKRD45 Tier 1 1 173846152 T C 1:173846152:T:C 2.56E-08 KLHL20 Tier 1 1 173846152 T C 1:173846152:T:C 2.56E-08 PRDX6 Tier 1 1 173846152 T C 1:173846152:T:C 2.56E-08 SERPINC1 Tier 1 1 173846152 T C 1:173846152:T:C 2.56E-08 SLC9C2 Tier 1 1 173846152 T C 1:173846152:T:C 2.56E-08 xxxxx CACNA1E Nearest 1 181676091 G A 1:181676091:G:A 8.85E-07 xxxxx VRK2 Nearest 2 57808420 G A 2:57808420:G:A 9.49E-07 xxxxx PLCL1 Nearest 2 197882813 A G 2:197882813:A:G 6.64E-07 xxxxx HTT Nearest 4 3240118 C T 4:3240118:C:T 8.03E-07 xxxxx ECI2 Nearest 6 4336259 T C 6:4336259:T:C 2.90E-07 xxxxx BTN2A2 Tier 1 6 26239176 A G 6:26239176:A:G 4.11E-09 TRIM38 Tier 1 6 26239176 A G 6:26239176:A:G 4.11E-09 ZNF322 Tier 1 6 26239176 A G 6:26239176:A:G 4.11E-09 ABT1 Tier 1 6 26239176 A G 6:26239176:A:G 4.11E-09 HFE Tier 1 6 26239176 A G 6:26239176:A:G 4.11E-09 BTN3A3 Tier 1 6 26239176 A G 6:26239176:A:G 4.11E-09 HMGN4 Tier 1 6 26239176 A G 6:26239176:A:G 4.11E-09 H4C8 MAGMA, Nearest 6 26239176 A G 6:26239176:A:G 4.11E-09 xxxxx ZNF311 MAGMA 6 29016371 G A 6:29016371:G:A 2.25E-06 xxxxx FBXL4 Tier 2 6 97984426 C CA 6:97984426:C:CA 4.85E-08 POU3F2 Nearest (1 of 2) 6 97984426 C CA 6:97984426:C:CA 4.85E-08 MMS22L Nearest (1 of 2) 6 97984426 C CA 6:97984426:C:CA 4.85E-08 xxxxx MLLT10 Nearest (1 of 2) 10 21748880 A G 10:21748880:A:G 6.34E-07 DNAJC1 Nearest (1 of 2) 10 21748880 A G 10:21748880:A:G 6.34E-07 xxxxx SOX6 Nearest 11 16217844 C G 11:16217844:C:G 1.08E-07 xxxxx SLC2A14 Nearest 12 7860921 T A 12:7860921:T:A 5.79E-07 xxxxx SUDS3 Tier 1, MAGMA 12 118202773 CTTTTTTTTTTTTT C 12:118202773:CTTTTTTTTTTTTT:C 1.64E-07 PEBP1 Tier 1 12 118202773 CTTTTTTTTTTTTT C 12:118202773:CTTTTTTTTTTTTT:C 1.64E-07 VSIG10 Tier 1 12 118202773 CTTTTTTTTTTTTT C 12:118202773:CTTTTTTTTTTTTT:C 1.64E-07 TAOK3 Nearest, MAGMA 12 118202773 CTTTTTTTTTTTTT C 12:118202773:CTTTTTTTTTTTTT:C 1.64E-07 xxxxx DNAH10 MAGMA 12 123924955 G A 12:123924955:G:A 2.43E-07 ZNF664 MAGMA 12 123924955 G A 12:123924955:G:A 2.43E-07 CCDC92 MAGMA 12 123924955 G A 12:123924955:G:A 2.43E-07 xxxxx OLFM4 Nearest, Tier 2 13 53194927 GT G 13:53194927:GT:G 1.16E-07 xxxxx PCDH17 Nearest 13 58456743 T C 13:58456743:T:C 9.42E-07 xxxxx CCPG1 Tier 2 15 54866724 A G 15:54866724:A:G 7.62E-09 UNC13C Nearest 15 54866724 A G 15:54866724:A:G 7.62E-09 xxxxx SHISA6 Nearest 17 11325637 G C 17:11325637:G:C 8.26E-08 xxxxx CA10 Tier 1, Nearest 17 52183006 C T 17:52183006:C:T 2.11E-09 xxxxx DCC Nearest 18 53232948 C T 18:53232948:C:T 2.48E-07 xxxxx CDK5RAP1 Nearest 20 33363039 G A 20:33363039:G:A 5.41E-07 xxxxx CSE1L Tier 1, MAGMA 20 48914387 T TA 20:48914387:T:TA 9.51E-12 ARFGEF2 Tier 1, MAGMA, Nearest 20 48914387 T TA 20:48914387:T:TA 9.51E-12 DDX27 Tier 1 20 48914387 T TA 20:48914387:T:TA 9.51E-12 STAU1 Tier 1, MAGMA 20 48914387 T TA 20:48914387:T:TA 9.51E-12 ZNFX1 Tier 1 20 48914387 T TA 20:48914387:T:TA 9.51E-12 B4GALT5 Tier 1 20 48914387 T TA 20:48914387:T:TA 9.51E-12 PTGIS Tier 1 20 48914387 T TA 20:48914387:T:TA 9.51E-12 xxxxx MRPL39 Nearest 21 25487862 A AG 21:25487862:A:AG 5.10E-07
LRRC7
NEGR1
LRRIQ3
ZNF644
RABGAP1L
DARS2
RC3H1
GPR52
ZBTB37
TNFSF4
ANKRD45
KLHL20
PRDX6
SERPINC1
SLC9C2
CACNA1E
VRK2
PLCL1
HTT
ECI2
BTN2A2
TRIM38
ZNF322
ABT1
HFE
BTN3A3
HMGN4
H4C8
ZNF311
FBXL4
POU3F2
MMS22L
MLLT10
DNAJC1
SOX6
SLC2A14
SUDS3
PEBP1
VSIG10
TAOK3
DNAH10
ZNF664
CCDC92
OLFM4
PCDH17
CCPG1
UNC13C
SHISA6
CA10
DCC
CDK5RAP1
CSE1L
ARFGEF2
DDX27
STAU1
ZNFX1
B4GALT5
PTGIS
MRPL39
NEGR1
LRRIQ3
ZNF644
RABGAP1L
DARS2
RC3H1
GPR52
ZBTB37
TNFSF4
ANKRD45
KLHL20
PRDX6
SERPINC1
SLC9C2
CACNA1E
VRK2
PLCL1
HTT
ECI2
BTN2A2
TRIM38
ZNF322
ABT1
HFE
BTN3A3
HMGN4
H4C8
ZNF311
FBXL4
POU3F2
MMS22L
MLLT10
DNAJC1
SOX6
SLC2A14
SUDS3
PEBP1
VSIG10
TAOK3
DNAH10
ZNF664
CCDC92
OLFM4
PCDH17
CCPG1
UNC13C
SHISA6
CA10
DCC
CDK5RAP1
CSE1L
ARFGEF2
DDX27
STAU1
ZNFX1
B4GALT5
PTGIS
MRPL39
Sources:
Tier 1 and 2 genes: Candidate genes document
MAGMA genes: Supplementary table 4
Top 25 loci: Supplementary table 3, with nearest gene to each lead variant found with LocusZoom. (Can also be done on UCSC Genome Browser.)
1. Go to genome browser at https://genome.ucsc.edu/cgi-bin/hgTracks
2. Press the "Hide All" button to turn off the unneeded tracks:
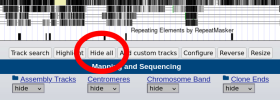
3. In the dropdown for GENCODE, select "pack", then press "Refresh":
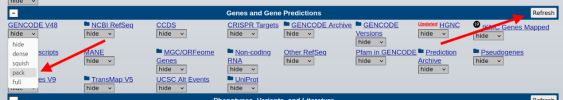
4. In the search box at the top, enter a chromosomal position in the format CHROMOSOME: POSITION, then press "Search". For example, 20:48914387 is the location of the most significant variant in DecodeME:
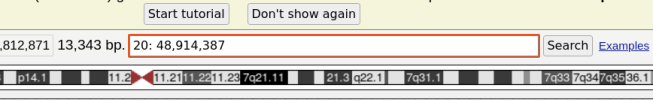
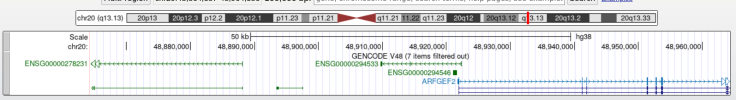
5. Press the buttons to the right of "Zoom out" at the top of the page until you see protein coding genes show up in the main field of view, which will be colored blue. For example, I had to press the 10x zoom out button five times to see that ARFGEF2 is the nearest gene to the most significant variant:
2. Press the "Hide All" button to turn off the unneeded tracks:

3. In the dropdown for GENCODE, select "pack", then press "Refresh":

4. In the search box at the top, enter a chromosomal position in the format CHROMOSOME: POSITION, then press "Search". For example, 20:48914387 is the location of the most significant variant in DecodeME:

5. Press the buttons to the right of "Zoom out" at the top of the page until you see protein coding genes show up in the main field of view, which will be colored blue. For example, I had to press the 10x zoom out button five times to see that ARFGEF2 is the nearest gene to the most significant variant:

Last edited:
I mainly compiled the list of candidate genes above to check if any of those genes are ranked highly for rare variant associations in the Genebass browser, which includes data from a study of all the phenotypes in the UK BioBank, including ME/CFS.
The rare variant study/dataset is discussed more here: Systematic single-variant and gene-based association testing of thousands of phenotypes in 394,841 UK Biobank exomes, 2022, Karczewski et al
So checking all the different p-values in Genebass (SKATO p, SKAT p, and burden p for missense, pLoF, and synonymous variant associations making 9 p-values per gene) for all 59 of the above genes, these are all the genes where the p-value was below .05 in any of the tests:
So just maybe there's sign of a real signal for VSIG10, but otherwise, the p-values aren't really low enough to give much confidence for these genes considering how many different gene-variant type associations are being considered (59 genes x 3 variant types = 177 tests. Three times as many, 531, if considering different statistical test types on the same gene as different tests, e.g. SKAT vs SKATO).
The rare variant study/dataset is discussed more here: Systematic single-variant and gene-based association testing of thousands of phenotypes in 394,841 UK Biobank exomes, 2022, Karczewski et al
So checking all the different p-values in Genebass (SKATO p, SKAT p, and burden p for missense, pLoF, and synonymous variant associations making 9 p-values per gene) for all 59 of the above genes, these are all the genes where the p-value was below .05 in any of the tests:
Gene P-value Variant type Statistical test VSIG10 2.53E-05 synonymous Burden VSIG10 3.38E-05 synonymous SKATO ANKRD45 0.00099 synonymous SKAT MMS22L 0.00109 pLoF SKAT MMS22L 0.00118 pLoF SKATO ANKRD45 0.00162 synonymous SKATO MMS22L 0.00450 pLoF Burden ANKRD45 0.00585 pLoF SKAT UNC13C 0.00914 synonymous Burden ANKRD45 0.01026 pLoF SKATO DCC 0.01077 pLoF SKAT SLC2A14 0.01204 missense|LC Burden DCC 0.01345 pLoF SKATO UNC13C 0.01723 synonymous SKATO SLC2A14 0.01848 missense|LC SKATO TAOK3 0.02115 pLoF SKAT CDK5RAP1 0.02116 synonymous SKAT CDK5RAP1 0.02325 synonymous SKATO PLCL1 0.02359 synonymous SKAT ANKRD45 0.02551 synonymous Burden VSIG10 0.02558 synonymous SKAT RABGAP1L 0.02674 pLoF SKAT ZBTB37 0.02692 pLoF SKAT TNFSF4 0.02708 missense|LC SKAT DCC 0.02809 pLoF Burden CDK5RAP1 0.02935 synonymous Burden PLCL1 0.03497 synonymous SKATO ZNF644 0.03584 synonymous Burden TAOK3 0.03714 pLoF SKATO SLC2A14 0.03919 missense|LC SKAT ZNF644 0.03954 synonymous SKAT VSIG10 0.03994 pLoF SKAT ZBTB37 0.04032 pLoF SKATO ZNF644 0.04152 synonymous SKATO TNFSF4 0.04163 synonymous SKAT SUDS3 0.04191 synonymous SKAT HTT 0.04427 pLoF SKAT RABGAP1L 0.04841 pLoF SKATO
So just maybe there's sign of a real signal for VSIG10, but otherwise, the p-values aren't really low enough to give much confidence for these genes considering how many different gene-variant type associations are being considered (59 genes x 3 variant types = 177 tests. Three times as many, 531, if considering different statistical test types on the same gene as different tests, e.g. SKAT vs SKATO).
Last edited:
Re-reading the preprint paper, this non-scientist is struggling with how imputation was done (pooled cases+controls, versus separately ... if cases have unusual haplotypes surely separate imputation avoids forcing imputation from, essentially, population haplotypes ...).
Is it possible to tell which variants/loci were imputed - can't immediately find it in the paper/supplementary? I am sort-of thinking an imputed variant/locus is a less-reliable signal than a chip-detected SNP.
Is it possible to tell which variants/loci were imputed - can't immediately find it in the paper/supplementary? I am sort-of thinking an imputed variant/locus is a less-reliable signal than a chip-detected SNP.
Kitty
Senior Member (Voting Rights)
@forestglip, would it be worth making a locked topic with your list of main candidate genes, which only gets added to if other candidates appear?
I've once or twice found myself reading things that might be related, then having to spend ages trying to remember whereabouts in a very long thread the list of genes last appeared. It would make it easier to find for anyone who's got time and energy to do a bit of digging.
Calling it something simple like 'Main candidate genes list 2025' would make it more searchable too. I always struggle to relocate topics about papers with long titles, as it's hard to remember one word that was definitely in it.
I've once or twice found myself reading things that might be related, then having to spend ages trying to remember whereabouts in a very long thread the list of genes last appeared. It would make it easier to find for anyone who's got time and energy to do a bit of digging.
Calling it something simple like 'Main candidate genes list 2025' would make it more searchable too. I always struggle to relocate topics about papers with long titles, as it's hard to remember one word that was definitely in it.
My shallow understanding is that imputation is done per person in a study, not pooling any participants together, by comparing that person's DNA to a whole genome reference panel, like 1000 Genomes. They look at the pattern of SNPs that they were actually able to test in a person, and see how that pattern compares to the whole genomes in the reference panel to see if a given pattern is associated with high confidence with other untested SNPs.Re-reading the preprint paper, this non-scientist is struggling with how imputation was done (pooled cases+controls, versus separately ... if cases have unusual haplotypes surely separate imputation avoids forcing imputation from, essentially, population haplotypes ...).
I wouldn't think it would cause issues for the reference panel to be healthy unlike the cases being imputed, but I'm not sure.
Whether SNPs were imputed in DecodeME is in the summary statistics files, where 1 means the SNP was actually measured, and anything less is a score for confidence about the imputation. For example:
Of the 8 hits, only 1 was measured, but the others have a high INFO_SCORE suggestion that their distributions follow Hardy-Weinberg Equilibrium.
View attachment 27867
I suppose it's based on the strong correlations between SNPs that you only need to know a few to be pretty certain what the others are. But I was quite surprised that the actually measured SNPs are so low (around 5% of the total, apparently).
You mean specifically genes that the DecodeME data suggests might be interesting or based on any source? Including any ME/CFS studies would be a lot more of a challenge depending on what sources are considered.Calling it something simple like 'Main candidate genes list 2025' would make it more searchable too. I always struggle to relocate topics about papers with long titles, as it's hard to remember one word that was definitely in it.
It might be useful. Some other options are linking to that gene list post from the first post of this thread, or bookmarking that post with the forum bookmark tool. What do you think? I'll also bring it up with the other mods.
Kitty
Senior Member (Voting Rights)
I just meant that list you made in post #793.You mean specifically genes that the DecodeME data suggests might be interesting or based on any source?
@Kitty, here is that thread: https://www.s4me.info/threads/main-candidate-genes-from-decodeme-2025.46949/
Whether SNPs were imputed in DecodeME is in the summary statistics files, where 1 means the SNP was actually measured,
Brilliant - I never would have found that.
For example, if ALL pathological ME variants are absent on the UKB Axiom array, how sure can we be that imputation against the general population will cause them to be associated with pwME?I wouldn't think it would cause issues for the reference panel to be healthy unlike the cases being imputed, but I'm not sure.
I don't think I understand imputation well enough to answer. In any case, we know there is a lot of the genome data missing in DecodeME. Imputation can only get you so far, and some of the imputed variants might be wrong.For example, if ALL pathological ME variants are absent on the UKB Axiom array, how sure can we be that imputation against the general population will cause them to be associated with pwME?
A whole genome sequencing study, AKA SequenceME, would be very valuable so that we could actually measure all the positions instead of making educated guesses about all the non-measured positions.
Jonathan Edwards
Senior Member (Voting Rights)
My impression is that 'imputation' is used to cover more than one inferential process, at least one involving individual SNP profiles (imputing intervening SNPs) and another making use of population weightings of SNP forms (imputing risk genes from minihaplotypes of SNPs).
On the TNFSF4 (also known as OX40L):
a paper on lupus (SLE) quoted by Forestglip said that an up-regulation of TNFSF4/OX40L (the OX40 ligand) predisposes to lupus. I think TNFSF4 activates CD4+ t-cells:
BUT DecodeME seems to be suggesting the opposite for ME/CFS - a decreased expression of TNFSF4, at least in some defined tissues
Members have noted that some drugs that reduce expression of TNFSF4 are coming to market and there was a suggestion that it could be tried in ME/CFS.
If I have understood things correctly, (and I may well not have, in which case, let me know), knocking down expression of TNFSF4 in people with ME/CFS would be very unlikely to help. The genetic variant found by DecodeME that reduces TNFSF4 expression might only be relevant to increasing the risk of developing ME/CFS, so an OXO40L monoclonal that reduces TNFSF4 expression might not make symptoms worse, but there doesn't seem to be a good rationale to try it.
?
a paper on lupus (SLE) quoted by Forestglip said that an up-regulation of TNFSF4/OX40L (the OX40 ligand) predisposes to lupus. I think TNFSF4 activates CD4+ t-cells:
We hypothesize that increased expression of TNFSF4 predisposes to SLE either by quantitatively augmenting T cell–APC interaction or by influencing the functional consequences of T cell activation via TNFRSF4.
Another quoted paper suggests that blocking the ligand and receptor can improve lupus:Two tumor necrosis factor (TNF) superfamily members located within intervals showing genetic linkage with SLE are TNFSF4 (also known as OX40L; 1q25), which is expressed on activated antigen-presenting cells (APCs)7,8 and vascular endothelial cells9, and also its unique receptor, TNFRSF4 (also known as OX40; 1p36), which is primarily expressed on activated CD4+ T cells10.
TNFSF4 produces a potent co-stimulatory signal for activated CD4+ T cells after engagement of TNFRSF4 (ref. 11).
Using both a family-based and a case-control study design, we show that the upstream region of TNFSF4 contains a single risk haplotype for SLE, which is correlated with increased expression of both cell-surface TNFSF4 and the TNFSF4 transcript.
Blockade of OX40/OX40L signaling using anti-OX40L alleviates murine lupus nephritis (2024)
BUT DecodeME seems to be suggesting the opposite for ME/CFS - a decreased expression of TNFSF4, at least in some defined tissues
The DecodeME paper says that the ME/CFS variants here are associated with decreased expression of TNFSF4 in the lung, skin of sun exposed lower leg, and thyroid.
Members have noted that some drugs that reduce expression of TNFSF4 are coming to market and there was a suggestion that it could be tried in ME/CFS.
A few OX40L monoclonals are coming to market soon:
This particular monoclonal is made by Sanofi, who recently agreed to let Scheibenbogen trial their CD38 inhibitor in ME/CFS. So if there is a rationale for trying their OX40 monoclonal in ME they might well be amenable.
If I have understood things correctly, (and I may well not have, in which case, let me know), knocking down expression of TNFSF4 in people with ME/CFS would be very unlikely to help. The genetic variant found by DecodeME that reduces TNFSF4 expression might only be relevant to increasing the risk of developing ME/CFS, so an OXO40L monoclonal that reduces TNFSF4 expression might not make symptoms worse, but there doesn't seem to be a good rationale to try it.
?
Last edited:
I think there are often situations where the same variant can be associated with increases or decreases of gene expression depending on the tissue, or maybe depending on other factors like age.The genetic variant found by DecodeME that reduces TNFSF4 expression might only be relevant to increasing the risk of developing ME/CFS, so it might not make symptoms worse, but there doesn't seem to be a good rationale to try it.
For example, a variant from a past study:
So the only significant finding in terms of genetic mutations was a SNP which affects the expression of CCK (aka cholecystokinin). The database they used seems to say that this SNP decreases expression of CCK in cultured fibroblasts but increases expression in colon cells.
Apart from that, the gene expression database isn't comprehensive of every tissue that might be affected, both because there might be too low of statistical power and because not every possible cell type/tissue type is included in the database.
So just noting that it's possible this variant might lead to increased expression where it matters for ME/CFS. Or of course you might be right and it could be a dead end.
V.R.T.
Senior Member (Voting Rights)
So I may be misremembering but the Sanofi OX40L monoclonal was claimed by the company to have a t cell calming/soothing effect, and JE and others have theorised T Cells may be central to the mechinism of MECFS. So I was going from that and the connections I saw iirc.If I have understood things correctly, (and I may well not have, in which case, let me know), knocking down expression of TNFSF4 in people with ME/CFS would be very unlikely to help. The genetic variant found by DecodeME that reduces TNFSF4 expression might only be relevant to increasing the risk of developing ME/CFS, so it might not make symptoms worse, but there doesn't seem to be a good rationale to try it.
?
But i didnt realise it was down not up in DecodeME.
Kitty
Senior Member (Voting Rights)
@Kitty, here is that thread: https://www.s4me.info/threads/main-candidate-genes-from-decodeme-2025.46949/
Thank you! Bookmarked.
I don't think I understand imputation well enough to answer.
I have spent an hour or two grappling with phasing and imputation, but realize the task is very large and complex. It just bothers me that most of the SNPs in the preprint are inferred by fiendish arithmetic.
As you say, SequenceME will be a blessing. Is there currently ANY whole genome data for ME? Is SequenceME the only WGS project on the horizon?
