When I previously ran LDSC to test for genetic correlations between DecodeME and all the traits in the UK Biobank, the correlation with lupus didn't work, likely because the sample size in the Biobank was too small.
Recent discussions about lupus made me want to see if I could get it working with a different dataset. I found a larger SLE study, Bentham 2015 [1], and the summary statistics for this study are downloadable from a couple locations, like Open GWAS. This website says the study includes 5201 cases and 9066 controls*.
I ran LDSC between the DecodeME data and the SLE data, again using Bigagwas, and here are the results:
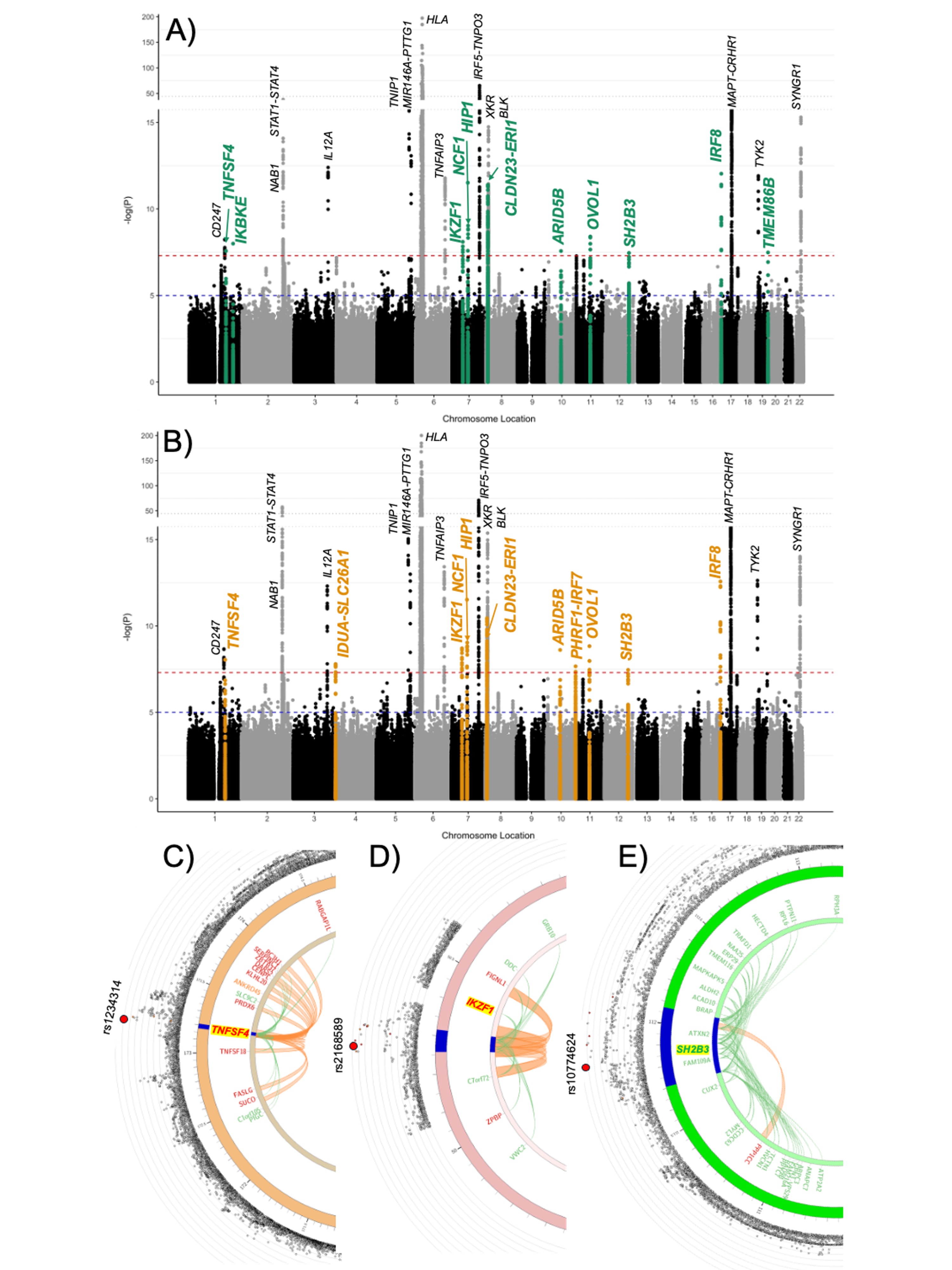
I thought it might be interesting to look at some of the significant regions to see how they compare between the two studies.
Here is that RABGAP1L region. SLE is in blue and ME/CFS is in red.

chr6p22.2:

Not much else is very significant for SLE around the regions of the rest of the top 8 ME/CFS loci.
1. Bentham, James et al. “Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus.” Nature genetics vol. 47,12 (2015): 1457-1464. doi:10.1038/ng.3434 https://pmc.ncbi.nlm.nih.gov/articles/PMC4668589/
Recent discussions about lupus made me want to see if I could get it working with a different dataset. I found a larger SLE study, Bentham 2015 [1], and the summary statistics for this study are downloadable from a couple locations, like Open GWAS. This website says the study includes 5201 cases and 9066 controls*.
I ran LDSC between the DecodeME data and the SLE data, again using Bigagwas, and here are the results:
So genetic correlation is 0.27, which is small but not nothing, and the correlation is significant at p=6.9e-7.Heritability of phenotype 1 [ME/CFS]
---------------------------
Total Liability scale h2: 0.0885 (0.0058)
Lambda GC: 1.1019
Mean Chi^2: 1.1428
Intercept: 0.9089 (0.0075)
Ratio < 0 (usually indicates GC correction).
Heritability of phenotype 2/2 [Systemic lupus erythematosus]
-----------------------------
Total Liability scale h2: 0.1789 (0.022)
Lambda GC: 1.4673
Mean Chi^2: 1.3551
Intercept: 1.2021 (0.0093)
Ratio: 0.5692 (0.0262)
Genetic Covariance
------------------
Total Liability scale gencov: 0.0334 (0.0071)
Mean z1*z2: 0.0341
Intercept: -0.0212 (0.0062)
Genetic Correlation
-------------------
Genetic Correlation: 0.2658 (0.0535)
Z-score: 4.9637
P: 6.9154e-07
I thought it might be interesting to look at some of the significant regions to see how they compare between the two studies.
Here is that RABGAP1L region. SLE is in blue and ME/CFS is in red.

chr6p22.2:

Not much else is very significant for SLE around the regions of the rest of the top 8 ME/CFS loci.
I think the numbers given for sample size on Open GWAS might be incorrect. When looking at the paper, the numbers given match with the sample size for the meta-analysis which included the main GWAS plus the Hom et al data.
But the p-values in the summary statistics seem to match the results from only the main GWAS when looking at Supplementary Table 3a. This would mean the sample size would be 4036 SLE cases and 6959 controls.
I used the larger sample size given by Open GWAS for LDSC, but this might not give exactly the right results if it should actually be the smaller sample size, which is about 25% smaller.
But the p-values in the summary statistics seem to match the results from only the main GWAS when looking at Supplementary Table 3a. This would mean the sample size would be 4036 SLE cases and 6959 controls.
I used the larger sample size given by Open GWAS for LDSC, but this might not give exactly the right results if it should actually be the smaller sample size, which is about 25% smaller.
1. Bentham, James et al. “Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus.” Nature genetics vol. 47,12 (2015): 1457-1464. doi:10.1038/ng.3434 https://pmc.ncbi.nlm.nih.gov/articles/PMC4668589/