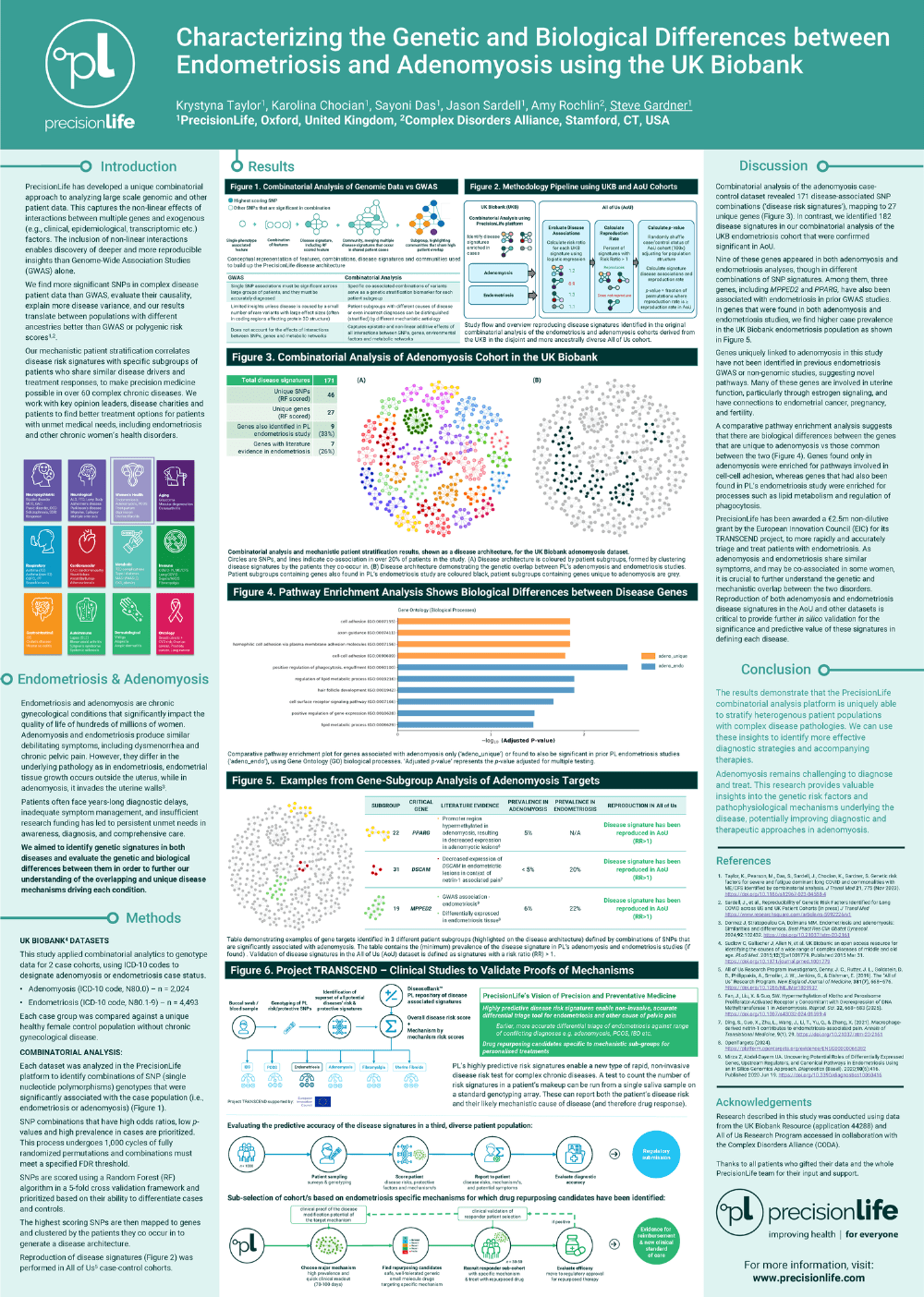
Identification and Validation of Novel Combinatorial Genetic Risk Factors for Endometriosis across Multiple UK and US Patient Cohorts
JM Sardell, S Das, GL Møller, M Sanna, K Chocian, K Taylor, AR Malinowski, C Stubberfield, A Rochlin, S Gardner
Background
Endometriosis affects about 10% of women usually of reproductive age. It often has severe negative impacts on patients’ quality of life, but the average time to a definitive diagnosis remains 7-9 years, and there are few effective therapeutic options. Relatively little is known about the genetic drivers of the disease even though heritability of the disease is fairly high. A recent large genome wide association study (GWAS) meta-analysis identified 42 genomic loci associated with risk of endometriosis, but together these explain only 5% of disease variance.
Methods
We used the PrecisionLife® combinatorial analytics platform to identify multi-SNP disease signatures significantly associated with endometriosis in a white European UK Biobank (UKB) cohort. We assessed the reproducibility of these multi-SNP disease signatures as well as 35 of the 42 SNPs identified by a recent meta-GWAS study in a multi-ancestry American endometriosis cohort from All of Us (AoU) after controlling for population structure.
Results
We identified 1,709 disease signatures, comprising 2,957 unique SNPs in combinations of 2-5 SNPs, that were associated with increased prevalence of endometriosis in UKB. We observed a significant enrichment of these signatures (58-88%, p<0.04) that are also positively associated with endometriosis in the AoU cohort, including one 2-SNP signature that is individually significant. Reproducibility rates were greatest for higher frequency signatures, ranging from 80-88% for signatures with greater than 9% frequency (p<0.01) in AoU. Encouragingly, the disease signatures also show high reproducibility rates in non-white European AoU sub-cohorts (66-76%, p<0.04 for signatures with greater than 4% frequency).
A total of 195 unique SNPs mapping to 100 genes were identified in the high frequency reproducing signatures (>9%). Of these, 4 genes were previously identified in the endometriosis meta-GWAS study and 19 genes have a previous association with endometriosis in OpenTargets1. 77 novel genes were identified in this study.
We characterized 9 novel genes that occur at the highest frequency in reproducing signatures and that do not contain any SNPs linked to known GWAS genes, providing new evidence for links between endometriosis and autophagy and macrophage biology. Reproducibility rates, ranging between 73% to 85%. are especially strong for the signatures that contain these 9 genes independently of any SNPs mapping to the meta-GWAS genes. These genes also include several targets novel to endometriosis with credible therapeutic discovery, repurposing and/or repositioning potential.
Conclusion
Although using much smaller, less well-characterized datasets than the previous whole genome meta-GWAS study, combinatorial analysis has provided important new insights into the genetics and biology of endometriosis. The finding of 77 novel gene associations that have high frequency and reproduce in an independent, ancestrally diverse dataset demonstrates that combinatorial analysis can identify biologically relevant genes that are overlooked by GWAS approaches. Several of these novel genes will are credible targets for drug discovery and repurposing, as shown by the examples highlighted.
The broad reproducibility of results across datasets and ancestries suggests that combinatorial disease signatures can be used to identify different mechanistic etiologies that have the potential to inform precision medicine-based approaches and generate new clinical treatments for this complex disease.
Web | PDF | Preprint: MedRxiv | Open Access
JM Sardell, S Das, GL Møller, M Sanna, K Chocian, K Taylor, AR Malinowski, C Stubberfield, A Rochlin, S Gardner
Background
Endometriosis affects about 10% of women usually of reproductive age. It often has severe negative impacts on patients’ quality of life, but the average time to a definitive diagnosis remains 7-9 years, and there are few effective therapeutic options. Relatively little is known about the genetic drivers of the disease even though heritability of the disease is fairly high. A recent large genome wide association study (GWAS) meta-analysis identified 42 genomic loci associated with risk of endometriosis, but together these explain only 5% of disease variance.
Methods
We used the PrecisionLife® combinatorial analytics platform to identify multi-SNP disease signatures significantly associated with endometriosis in a white European UK Biobank (UKB) cohort. We assessed the reproducibility of these multi-SNP disease signatures as well as 35 of the 42 SNPs identified by a recent meta-GWAS study in a multi-ancestry American endometriosis cohort from All of Us (AoU) after controlling for population structure.
Results
We identified 1,709 disease signatures, comprising 2,957 unique SNPs in combinations of 2-5 SNPs, that were associated with increased prevalence of endometriosis in UKB. We observed a significant enrichment of these signatures (58-88%, p<0.04) that are also positively associated with endometriosis in the AoU cohort, including one 2-SNP signature that is individually significant. Reproducibility rates were greatest for higher frequency signatures, ranging from 80-88% for signatures with greater than 9% frequency (p<0.01) in AoU. Encouragingly, the disease signatures also show high reproducibility rates in non-white European AoU sub-cohorts (66-76%, p<0.04 for signatures with greater than 4% frequency).
A total of 195 unique SNPs mapping to 100 genes were identified in the high frequency reproducing signatures (>9%). Of these, 4 genes were previously identified in the endometriosis meta-GWAS study and 19 genes have a previous association with endometriosis in OpenTargets1. 77 novel genes were identified in this study.
We characterized 9 novel genes that occur at the highest frequency in reproducing signatures and that do not contain any SNPs linked to known GWAS genes, providing new evidence for links between endometriosis and autophagy and macrophage biology. Reproducibility rates, ranging between 73% to 85%. are especially strong for the signatures that contain these 9 genes independently of any SNPs mapping to the meta-GWAS genes. These genes also include several targets novel to endometriosis with credible therapeutic discovery, repurposing and/or repositioning potential.
Conclusion
Although using much smaller, less well-characterized datasets than the previous whole genome meta-GWAS study, combinatorial analysis has provided important new insights into the genetics and biology of endometriosis. The finding of 77 novel gene associations that have high frequency and reproduce in an independent, ancestrally diverse dataset demonstrates that combinatorial analysis can identify biologically relevant genes that are overlooked by GWAS approaches. Several of these novel genes will are credible targets for drug discovery and repurposing, as shown by the examples highlighted.
The broad reproducibility of results across datasets and ancestries suggests that combinatorial disease signatures can be used to identify different mechanistic etiologies that have the potential to inform precision medicine-based approaches and generate new clinical treatments for this complex disease.
Web | PDF | Preprint: MedRxiv | Open Access