wigglethemouse
Senior Member (Voting Rights)
Genome-wide association study of multisite chronic pain in UK Biobank, 2019, Johnston
Abstract
Chronic pain is highly prevalent worldwide and represents a significant socioeconomic and public health burden. Several aspects of chronic pain, for example back pain and a severity-related phenotype ‘chronic pain grade’, have been shown previously to be complex heritable traits with a polygenic component. Additional pain-related phenotypes capturing aspects of an individual’s overall sensitivity to experiencing and reporting chronic pain have also been suggested as a focus for investigation.
We made use of a measure of the number of sites of chronic pain in individuals within the UK general population. This measure, termed Multisite Chronic Pain (MCP), is a complex trait and its genetic architecture has not previously been investigated. To address this, we carried out a large-scale genome-wide association study (GWAS) of MCP in ~380,000 UK Biobank participants.
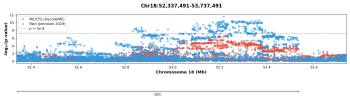
Our findings were consistent with MCP having a significant polygenic component, with a Single Nucleotide Polymorphism (SNP) heritability of 10.2%. In total 76 independent lead SNPs at 39 risk loci were associated with MCP.
Additional gene-level association analyses identified neurogenesis, synaptic plasticity, nervous system development, cell-cycle progression and apoptosis genes as enriched for genetic association with MCP. Genetic correlations were observed between MCP and a range of psychiatric, autoimmune and anthropometric traits, including major depressive disorder (MDD), asthma and Body Mass Index (BMI).
Furthermore, in Mendelian randomisation (MR) analyses a causal effect of MCP on MDD was observed. Additionally, a polygenic risk score (PRS) for MCP was found to significantly predict chronic widespread pain (pain all over the body), indicating the existence of genetic variants contributing to both of these pain phenotypes.
Overall, our findings support the proposition that chronic pain involves a strong nervous system component with implications for our understanding of the physiology of chronic pain. These discoveries may also inform the future development of novel treatment approaches.
LINK | PDF | PLOS Genetics
Abstract
Chronic pain is highly prevalent worldwide and represents a significant socioeconomic and public health burden. Several aspects of chronic pain, for example back pain and a severity-related phenotype ‘chronic pain grade’, have been shown previously to be complex heritable traits with a polygenic component. Additional pain-related phenotypes capturing aspects of an individual’s overall sensitivity to experiencing and reporting chronic pain have also been suggested as a focus for investigation.
We made use of a measure of the number of sites of chronic pain in individuals within the UK general population. This measure, termed Multisite Chronic Pain (MCP), is a complex trait and its genetic architecture has not previously been investigated. To address this, we carried out a large-scale genome-wide association study (GWAS) of MCP in ~380,000 UK Biobank participants.
Our findings were consistent with MCP having a significant polygenic component, with a Single Nucleotide Polymorphism (SNP) heritability of 10.2%. In total 76 independent lead SNPs at 39 risk loci were associated with MCP.
Additional gene-level association analyses identified neurogenesis, synaptic plasticity, nervous system development, cell-cycle progression and apoptosis genes as enriched for genetic association with MCP. Genetic correlations were observed between MCP and a range of psychiatric, autoimmune and anthropometric traits, including major depressive disorder (MDD), asthma and Body Mass Index (BMI).
Furthermore, in Mendelian randomisation (MR) analyses a causal effect of MCP on MDD was observed. Additionally, a polygenic risk score (PRS) for MCP was found to significantly predict chronic widespread pain (pain all over the body), indicating the existence of genetic variants contributing to both of these pain phenotypes.
Overall, our findings support the proposition that chronic pain involves a strong nervous system component with implications for our understanding of the physiology of chronic pain. These discoveries may also inform the future development of novel treatment approaches.
LINK | PDF | PLOS Genetics
Last edited: