Gene expression profiles
See
Fig. 9: Male and female cohorts have distinct differential gene expression profiles in the muscle.
The title claims that male and female cohorts are different when it comes to gene expression in muscle. I could believe that. But look at the PCA charts:
View attachment 21163
Figure a
There's no overall difference between gene expression in the muscle between the healthy volunteers and ME/CFS group. They've used Principal Component 1 and Principal Component 2 - which explain 16.1% and 10.6% of the variation, which is not great. There's no clear difference between the two groups. Also, note the numbers of data points - I make that 8 healthy controls, 14 ME/CFS - also not great.
Figure b
And then, they give us Chart B, where all the males (healthy + ME/CFS = approx 11) are grouped together, and all the females from the two cohorts (approx 12) are grouped together. They claim that this shows that gene expression in muscle is different in males and females. But, first off, look at the separation of the male and female data points - yeah, there isn't anything like two clear groups. And then look at the axes labels. They are showing PC2 and PC3, explaining 10.6% and 9.7% of the variation. It's a pretty weak result to warrant that Figure title claiming differential gene expression in the muscle of males and females, particularly given that small sample size.
(BTW: Watch out for the use of PCAs that don't explain much variance elsewhere in the paper)
View attachment 21164
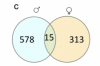
Figure c is a Venn diagram, I think showing that males had 593 differentially expressed genes (i.e. there is different expression between ME/CFS males and healthy males) in muscles, females had 328 differentially expressed genes in muscles, and, of those genes, only 15 genes were differentially expressed in both males and females. Given the tiny sample size (e.g. about 6 ME/CFS males and 6 healthy males) and the very poor replication of findings in the male and female subsamples, surely there are a lot of false positives there. While there may be some sex differences, I'm always dubious when a difference can only found by stratifying small samples into miniscule ones.
I think Figures d, e, f and g are mostly the product of the false positives and can be ignored. The authors take the expression values for each participant for only the genes that were differentially expressed between the ME/CFS and healthy cohorts. And then they do a Principal Component Analysis on them. And, amazing! the ME/CFS and healthy cohorts form discrete groups. Well, not so amazing, because they only used the data points that were different between the two groups, and didn't use the datapoints that didn't show a difference. So, those PCAs aren't really telling us anything new.
( Just further on that - the paper says "In the male PCA of 593 DE genes, clustering was observed based on the disease status (Fig.
9d, e)." and
"In the females, PCA of the 328 DE genes showed distinct clusters of HV and PI-ME/CFS samples (Fig.
9f, g)."
I think that's highly misleading and circular for the reason I explained in the preceding paragraph.)
Figures h,i, j, k - h and i are for males; j and k are for females.
The authors try to group the differentially expressed genes into pathways. The two separate figures for each sex are for pathways that are claimed to be up-regulated and down-regulated.
So, that's adding quite a layer of interpretation that may not be well founded.
@DMissa had some concerns about this approach - I've copied his comment here:
I couldn't see much overlap in the pathways listed for males and females, and find that worrying. Yes, there probably are sex differences, but males and females with ME/CFS both report that dead, heavy feeling in muscles in response to exertion, so there should be some commonality.
So...
there could be some gene expression differences in there that are of interest. In particular, I'd like to look at the 15 gene expressions that were different between ME/CFS and healthy volunteers in
both males and females.
But real gene expression differences are surely swamped by the false positives created by the small sample sizes and mis-matched cohorts.
(I think there is a stray sentence at the end of the Figure 9 caption that is actually talking about a supplementary figure.)
I'm done for the night. Feel free to point out errors or disagree with me.