The researchers compared fibroblasts
of S1 with her brother S2, who also carried a P38 mutation.
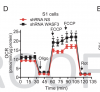
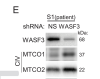
They found that S1's fibroblasts had lower oxygen consumption rate (OCR) and decreased Complex IV (one of the ion transport complexes in the mitochondrial membrane). Complex IV has two proteins MTCO1 and MTCO2. They found a 34% decrease in S1's MTCO1 compared to her brother.
The researchers found that S1's p53 was highly phosphorylated at a specific place, Ser46 (unlike her brother, who did not have the exertion intolerance etc symptoms). They knew that activated P38 MAPK14 could increase that phosphorylation, and indeed they found higher levels of that activated P38 MAPK14. WASF3 was known to activate P38 MAPK14, and there was the report of WASF3 being highly expressed in people with chronic fatigue syndrome. So, they looked at WASF3 levels in S1, and, sure enough, she had higher levels than her brother - 40% higher levels.
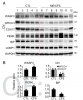
Jumping over the mice stuff for the moment, here's the finding on the level of WASF3 protein in 10 controls and 14 people with ME/CFS from the NIH ME/CFS study. So, WASF3 is also higher in the people with ME/CFS. There also seems to be less MTCO1.
View attachment 20147