jnmaciuch
Senior Member (Voting Rights)
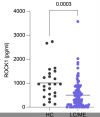
It is automatic in a couple softwares—this looks like GraphPrism, which is a very “user friendly” (meaning you don’t need to know a coding language to use it) propriety plotting software. It’s possible the user doesn’t even have access to a setting that would turn it off.Does anyone know, does that happen automatically in some widely used chart-making software, or do you have to select some options to consciously produce a chart like that?
[Added: The points in that plot appear to be automatically plotted by relative density, meaning that the sections of y axis with the highest density of points get plotted out horizontally to span the maximum width of the box plot, and then other “bins” with lower densities of points are spread out relative to that maximum]
Last edited: