jnmaciuch
Senior Member (Voting Rights)
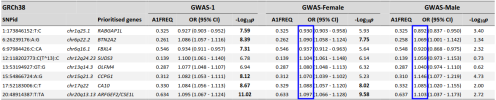
The document Chris just shared states that the variants were associated with decreased expression of BTN2A2.So if the gene inhibits the proliferation of CD4+CD8 t cells activated by IFNG does that provide evidence for or againsts JE et als hypothesis?
Inhibition would suggest against to me but I know little about these things.