Hoopoe
Senior Member (Voting Rights)
@Chris Ponting thank you for your work and dedication.
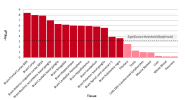
Ah yes apologies. I think I'm a bit confused because those low p-values shown in the graph are in purple and for the category: 0.01 ≤ MAF < 0.04, but I thought that the hits found had MAF > 10%?Why do you say that? The top hits on chr20 were around that p-value.
Oh good point. If I'm understanding correctly, the furthest right, uppermost dot is 20:48914387:T:TA (the lead hit in the whole study), but its MAF is around .34.Ah yes apologies. I think I'm a bit confused because those low p-values shown in the graph are in purple and for the category: 0.01 ≤ MAF < 0.04, but I thought that the hits found had MAF > 10%?
Hi Chris, sincere thanks for all your hard work and advocacy on our behalf. Exciting to hear more analyses of the data are in the works!Hi. Yes, we needed to LiftOver DNA variants from human genome build 38 (hg38) to the previous version (GRCh37 (hg19)) which FUMA uses for, e.g., MAGMA analysis. You're correct that some SNPs are lost in the LiftOver, but only a few. More variants are lost because FUMA's reference panel of variants is incomplete. Yes, the GWAS model we used in REGENIE is an additive linear model. But we are also applying non-additive, non-linear models separately (TarGene; will report when we have results). Yes - as with nearly all GWAS - are lead SNPs are not in protein-coding sequence. And, yes, there is scope for a lot more analyses both by us (DecodeME) and any researchers who receive the consented and pseudoanonymised data via the Data Access Committee. The preprint's title starts with "Initial Results..." and we mean this. Yet more to come.
No timeframe for completion of the HLA analysis. The Manhattan plot in the preprint includes all variants including those in the HLA. But note that in the HLA analyses we do not test each DNA variant, rather we test combinations of variants that are commonly coinherited, i.e. "HLA alleles". So even if there isn't a "signal" in the Manhattan plot this does not immediately mean that we won't see association to an HLA allele.Hi Chris, sincere thanks for all your hard work and advocacy on our behalf. Exciting to hear more analyses of the data are in the works!
I was wondering if there was any sort of timeframe for the publication of the HLA analysis? And am curious whether the Manhattan plot published with the initial data shows the HLA data points or whether they were removed from the plot whilst the analysis is ongoing?
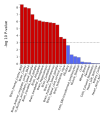
Tried to recreate the plot using the whole dataset:Don't think we have discussed this plot yet:

Looks like a labelling issue of the graph. Here's what I got trying to recreate it. The high p-values belong to the SNPs with high MAF values, showing that it was mostly this group that reached significant hits.Oh good point. If I'm understanding correctly, the furthest right, uppermost dot is 20:48914387:T:TA (the lead hit in the whole study), but its MAF is around .34.

awk -v OFS='\t' 'FNR==NR {ids[$1]++; next} NR==1 || ($3 in ids)' gwas_qced.var gwas_1.regenie > filtered_regenie_file.txtawk -v OFS='\t' 'NR!=1 { print "chr"$1, $2-1, $2, $0 }' filtered_regenie_file.txt > query.bed
sort-bed query.bed > sorted_query.bedzcat snp151.txt.gz | awk -v OFS='\t' '{if($12=="single") print $2,$3,$4,$5}' > snp151_snp.bedbedmap --echo --echo-map-id --delim '\t' sorted_query.bed snp151_snp.bed > sorted_query_with_rsids.bedawk -v OFS='\t' '$21 != "" {print $21,$4,$5}' sorted_query_with_rsids.bed > decodeme.magma_input.id.chr.genpos.txtawk -v OFS='\t' '$21 != "" {print $21,10^(-$19)}' sorted_query_with_rsids.bed > decodeme.magma_input.id.p.txtsnploc=./decodeme.magma_input.id.chr.genpos.txt
ncbi38=./NCBI38.gene.loc
magma --annotate \
--snp-loc ${snploc} \
--gene-loc ${ncbi38} \
--out decodemeref=./g1000_eur/g1000_eur
magma \
--bfile $ref \
--pval ./decodeme.magma_input.id.p.txt N=275488 \
--gene-annot decodeme.genes.annot \
--out decodemegeneset=msigdb_v2025.1.Hs_GMTs/c5.all.v2025.1.Hs.entrez.gmt
magma \
--gene-results decodeme.genes.raw \
--set-annot ${geneset} \
--out decodemeThanks for delving into this!As mentioned above, something @forestglip and I have been working on recently is running MAGMA locally on the DecodeME summary statistics.
I wouldn’t necessarily recommend anyone do this or follow these instructions. There may be things wrong with this method and it seems to end up with only 94% of variants mapped while @forestglip gets over 99% using the LiftOver and FUMA method mentioned earlier.
Unfortunately a lot of bioinformatics pipelines are pretty hodgepodge and you need to be a Jack of all trades to get something new up and running. It’s true that all of coding is 90% troubleshooting, but for bioinformatics especially where almost none of the packages/tools were designed by people explicitly trained in software engineering, it can be hell in a handbasket with entire weeks wasted. I started out in research with only a little bit of self-taught R experience and immediately got thrown headfirst into trying to understand what awk was with no help except stack exchange—I’m glad I had already developed the ability to parse through complete gibberish and not get overwhelmed, otherwise it would have discouraged me from science all together.Thanks for delving into this!
It looks so complicated. I expected that there would be nice packages in R and Python that wrap this into a handy user interface and provide the correct reference data (for LD, for example) automatically. Quite disappointing that the workflow mostly consists of command-line instructions and extracting the right data and tools from the web.
Yesterday I tried doing the LDSC and stumbled into one problem after the other (it only runs on Python2, need to convert GRCh38 to rsID, download the correct reference data online, etc.). I ran out of energy but hope to try again later.
Note that the script hotblack posted includes a lot of extras for convenience, like remembering settings for future runs. These are the only required steps we used:It looks so complicated. I expected that there would be nice packages in R and Python that wrap this into a handy user interface and provide the correct reference data (for LD, for example) automatically.
# First store all variants from gwas_qced.var in ids array.
# Then loop over every row in gwas_1.regenie and save to filtered_regenie_file.txt if it's the first row (header) or if the third column (the ID) is in the ids array.
awk 'FNR==NR {ids[$1]++; next} NR==1 || ($3 in ids)' gwas_qced.var gwas_1.regenie > filtered_regenie_file.txt
# For all but first row, print the chromosome, position-1, position, and entire original line to query.bed
# Need to subtract one because .bed files are zero indexed
awk 'NR!=1 { print "chr"$1, $2-1, $2, $0 }' filtered_regenie_file.txt > query.bed
# Sort bed file
sort-bed query.bed > sorted_query.bed
# Filter rsid reference file to only include SNVs.
# Pipe decompressed file to awk
# Set output separator to tab
# If the twelfth column is "single" (SNV), output chr, pos, pos, rsid (bed format)
zcat snp151.txt.gz | awk -v OFS='\t' '{if($12=="single") print $2,$3,$4,$5}' > snp151_snp.bed
# Add matching rs ids to the positions in the summary stats bed file
bedmap --echo --echo-map-id --delim '\t' sorted_query.bed snp151_snp.bed > sorted_query_with_rsids.bed
# Create SNP position file from summary statistics for MAGMA
# The format is 'rsid chromosome position'
awk '$21 != "" {print $21,$4,$5}' sorted_query_with_rsids.bed > decodeme.magma_input.id.chr.genpos.txt
# Create SNP p value file from summary statistics for MAGMA
# The format is 'rsid p-value'
awk '$21 != "" {print $21,10^(-$19)}' sorted_query_with_rsids.bed > decodeme.magma_input.id.p.txt
# Annotate genes with SNPs
snploc=./decodeme.magma_input.id.chr.genpos.txt
ncbi38=./NCBI38.gene.loc
magma --annotate \
--snp-loc ${snploc} \
--gene-loc ${ncbi38} \
--out decodeme
# Do gene-based analysis to create score for each gene
ref=./g1000_eur/g1000_eur
magma \
--bfile $ref \
--pval ./decodeme.magma_input.id.p.txt N=275488 \
--gene-annot decodeme.genes.annot \
--out decodeme
# Run gene-set analysis
geneset=msigdb_v2025.1.Hs_GMTs/c5.all.v2025.1.Hs.entrez.gmt
magma \
--gene-results decodeme.genes.raw \
--set-annot ${geneset} \
--out decodemeIt might not help with the practical problems—hard to know because I don't understand any of it!—but there is a catalogue of GWAS studies at the Biometrics Institute.The main struggle seems to be either mapping the variants to rs ids or converting from GRCh38 to GRCh37, and it doesn't seem that either is a trivial task to do exactly right. I'm not sure we did the rs id mapping perfectly since we based it only on positions and not letters. Surprisingly hard to find good explanations.
I’ve gained a whole new level of appreciation for what you and other people in bioinformatics do.…Unfortunately a lot of bioinformatics pipelines are pretty hodgepodge and you need to be a Jack of all trades to get something new up and running.
I appreciate it, though I’ll be honest it’s partially a mess of our own making. Not a month goes by where I don’t witness a PhD bioinformaticist doing something utterly baffling like hardcoding 16 separate subsets of one data table instead of creating one variable to split the table by…(and I wish that was an exaggeration not a real example)I’ve gained a whole new level of appreciation for what you and other people in bioinformatics do.…
Recently, integrative methods have been developed that aim to combine many different levels of functional data to predict the effector gene in a GWAS locus1–4. There are two main strategies to do so. The first strategy prioritizes genes using locus-based SNP-to-gene data. Examples of this are chromatin interaction mapping, quantitative trait loci (QTLs) mapping or selecting the closest gene to the lead variant. These annotations can then be combined by linear regression or machine learning to merge several types of SNP-to-gene data into a single prediction1,3. The second strategy assumes that all GWAS signal converges into shared, underlying biological pathways and networks. These methods prioritize genes in a locus based on gene features enriched across the entire GWAS4. However, no current method leverages these two strategies together to make well-calibrated predictions of the effector genes in a locus. We designed a new framework, called FLAMES. This framework integrates SNP-to-gene evidence and convergence-based evidence into a single prediction for each fine-mapped GWAS signal.

In our benchmarks, we found that the only competitive method to FLAMES is selecting the closest gene to lead SNP if it also has the highest PoPS in the locus. Generally, this produces slightly higher precision (3.1% across all benchmarks) than FLAMES would yield, without the need for running the FLAMES annotation and scoring steps, reducing the computational complexity of gene prioritization. However, prioritizing genes using FLAMES at the recommended threshold will yield approximately 33.2% more correctly identified causal genes.
Looks interesting! Not sure what sort of computing power it would require…I've noticed that the Dutch authors of MAGMA have a new method called FLAME
Looks interesting. I tried to see if I could do anything, but it's too much stuff I don't know how to do, like the part about creating credible set files.For those with strength, courage and coding skills:
I've noticed that the Dutch authors of MAGMA have a new method called FLAME. It combines multiple approaches to finding the effector gene within a significant GWAS locus using a machine-learning framework.
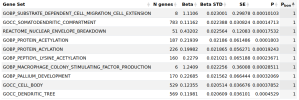
GOBP_SUBSTRATE_DEPENDENT_CELL_MIGRATION_CELL_EXTENSION
GOCC_SOMATODENDRITIC_COMPARTMENT
REACTOME_NUCLEAR_ENVELOPE_BREAKDOWN - Can't find a page for this
GOBP_PROTEIN_ACETYLATION
GOBP_PROTEIN_ACYLATION
GOBP_PEPTIDYL_LYSINE_ACETYLATION
GOBP_MACROPHAGE_COLONY_STIMULATING_FACTOR_PRODUCTION
GOBP_PALLIUM_DEVELOPMENT
GOCC_CELL_BODY
GOCC_DENDRITIC_TREE



