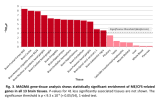
For members that are interested in exploring the data from DecodeME, such as to see if certain genes are near significant loci, here is a link to the summary stats on LocusZoom: https://my.locuszoom.org/gwas/894183/
I wasn't sure if I could share it publicly, but Chris Ponting kindly pointed out that the summary stats are released under a CC-By Attribution 4.0 International license, so sharing is allowed.
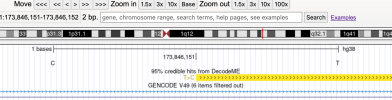
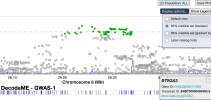
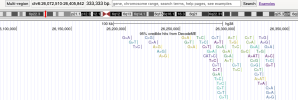
As an example of how to use LocusZoom to look at a gene:
I wasn't sure if I could share it publicly, but Chris Ponting kindly pointed out that the summary stats are released under a CC-By Attribution 4.0 International license, so sharing is allowed.
As an example of how to use LocusZoom to look at a gene:
- Find the location of a gene of interest. One way is to go the GeneCards page for a gene, scroll down to the section that says "Genomic Locations for ... Latest assembly", and copy the location which looks like this: 'chr6:31,575,565-31,578,336'
- Click the link that says "region page" on the LocusZoom page above. Paste the gene location you copied into the search box and press Enter, and it'll take you to the gene.
- You can zoom in or out by pressing Shift while 'scrolling' (e.g. drag two fingers on laptop touch pad or spin wheel on mouse).
Download
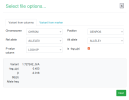
The script makes a file called gwas_1_sorted.txt.gz, which you would upload to LocusZoom. These are the options I set while uploading:
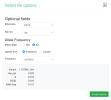
gwas_1.regenie.gz and gwas_qced.var.gz from OSF, run this code in a terminal in the same folder as the files to make a file suitable for LocusZoom, then upload to LocusZoom and set options as indicated in screenshots.
Bash:
# Filters to only QCed variants, removes non-needed columns, then sorts. Deletes intermediate files at the end.
summary_stats_file='./gwas_1.regenie.gz'
qced_var_file='./gwas_qced.var.gz'
awk 'FNR==NR {if (FNR>1) ids[$1]++; next} FNR==1 || ($3 in ids)' <(zcat "$qced_var_file") <(zcat "$summary_stats_file") > gwas_1_filtered.txt
awk 'BEGIN { OFS = "\t" } {print $1, $2, $4, $5, $6, $13, $14, $16}' gwas_1_filtered.txt > gwas_1_minimal.txt
awk 'NR==1; NR>1 {print $0 | "sort -k1,1n -k2,2n"}' gwas_1_minimal.txt | gzip > gwas_1_sorted.txt.gz
rm gwas_1_filtered.txt gwas_1_minimal.txtThe script makes a file called gwas_1_sorted.txt.gz, which you would upload to LocusZoom. These are the options I set while uploading:


Last edited: