Ekua W. Brenu ,Kevin J. Ashton,Jana Batovska,Donald R. Staines,Sonya M. Marshall-Gradisnik
Abstract
Background
MicroRNAs (miRNAs) are known to regulate many biological processes and their dysregulation has been associated with a variety of diseases including Chronic Fatigue Syndrome/Myalgic Encephalomyelitis (CFS/ME). The recent discovery of stable and reproducible miRNA in plasma has raised the possibility that circulating miRNAs may serve as novel diagnostic markers. The objective of this study was to determine the role of plasma miRNA in CFS/ME.
Results
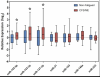
Using Illumina high-throughput sequencing we identified 19 miRNAs that were differentially expressed in the plasma of CFS/ME patients in comparison to non-fatigued controls. Following RT-qPCR analysis, we were able to confirm the significant up-regulation of three miRNAs (hsa-miR-127-3p, hsa-miR-142-5p and hsa-miR-143-3p) in the CFS/ME patients.
Conclusion
Our study is the first to identify circulating miRNAs from CFS/ME patients and also to confirm three differentially expressed circulating miRNAs in CFS/ME patients, providing a basis for further study to find useful CFS/ME biomarkers.
https://pubmed.ncbi.nlm.nih.gov/25238588/
Abstract
Background
MicroRNAs (miRNAs) are known to regulate many biological processes and their dysregulation has been associated with a variety of diseases including Chronic Fatigue Syndrome/Myalgic Encephalomyelitis (CFS/ME). The recent discovery of stable and reproducible miRNA in plasma has raised the possibility that circulating miRNAs may serve as novel diagnostic markers. The objective of this study was to determine the role of plasma miRNA in CFS/ME.
Results
Using Illumina high-throughput sequencing we identified 19 miRNAs that were differentially expressed in the plasma of CFS/ME patients in comparison to non-fatigued controls. Following RT-qPCR analysis, we were able to confirm the significant up-regulation of three miRNAs (hsa-miR-127-3p, hsa-miR-142-5p and hsa-miR-143-3p) in the CFS/ME patients.
Conclusion
Our study is the first to identify circulating miRNAs from CFS/ME patients and also to confirm three differentially expressed circulating miRNAs in CFS/ME patients, providing a basis for further study to find useful CFS/ME biomarkers.
https://pubmed.ncbi.nlm.nih.gov/25238588/