ME/CFS Science Blog
Senior Member (Voting Rights)
Of the 8 hits, only 1 was measured, but the others have a high INFO_SCORE suggestion that their distributions follow Hardy-Weinberg Equilibrium.
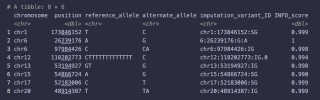

I suppose it's based on the strong correlations between SNPs that you only need to know a few to be pretty certain what the others are. But I was quite surprised that the actually measured SNPs are so low (around 5% of the total, apparently).Yes, I don't understand how that works.

