Nightsong
Senior Member (Voting Rights)
Abstract:
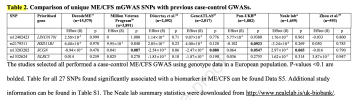
Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) is a clinically heterogeneous disease lacking approved therapies. To assess genetic susceptibility towards a specific metabolic phenotype, we performed a genome-wide association study on plasma biomarker levels (mGWAS) in ME/CFS patients (n=875) and healthy controls (HCs) (n=36,033). We identified 112 significant SNP–biomarker associations in ME/CFS, compared with 4,114 in HCs.
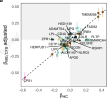
Two SNPs specific to ME/CFS, mapping to HSD11B1 and SCGN, were associated to phospholipids in extra-large very low-density lipoproteins (VLDL) and total fatty acids respectively. Genetic effects of VLDL associations were among the least correlated between ME/CFS and HCs. Heterogeneity tests found differential effects for several lipid traits at ADAP1, NR1H3 and CD40, which are involved in immune regulation. ME/CFS mGWAS summary statistics were decomposed to uncover shared genetic-metabolic patterns, where enrichment analysis highlighted pathways in lipid metabolism, neurotransmitter transport, and inflammation. These findings provide a genetic and molecular rationale for patient heterogeneity and suggest a polygenic predisposition in which many small-effect variants may jointly perturb metabolic mechanisms.
Link | PDF (iScience, December 2025, open access)
Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) is a clinically heterogeneous disease lacking approved therapies. To assess genetic susceptibility towards a specific metabolic phenotype, we performed a genome-wide association study on plasma biomarker levels (mGWAS) in ME/CFS patients (n=875) and healthy controls (HCs) (n=36,033). We identified 112 significant SNP–biomarker associations in ME/CFS, compared with 4,114 in HCs.
Two SNPs specific to ME/CFS, mapping to HSD11B1 and SCGN, were associated to phospholipids in extra-large very low-density lipoproteins (VLDL) and total fatty acids respectively. Genetic effects of VLDL associations were among the least correlated between ME/CFS and HCs. Heterogeneity tests found differential effects for several lipid traits at ADAP1, NR1H3 and CD40, which are involved in immune regulation. ME/CFS mGWAS summary statistics were decomposed to uncover shared genetic-metabolic patterns, where enrichment analysis highlighted pathways in lipid metabolism, neurotransmitter transport, and inflammation. These findings provide a genetic and molecular rationale for patient heterogeneity and suggest a polygenic predisposition in which many small-effect variants may jointly perturb metabolic mechanisms.
Link | PDF (iScience, December 2025, open access)
Last edited by a moderator: