Sly Saint
Senior Member (Voting Rights)
The Independent:
Gut bacteria could help diagnose long Covid and chronic fatigue syndrome, researchers find
Bacteria in the gut could help diagnose long Covid and chronic fatigue syndrome, researchers have found.
The debilitating condition, which can cause extreme tiredness, sleep problems, dizziness and brain fog, is often overlooked as there is no specific test to diagnose it. This means doctors have to simply rule out other illnesses.
But research published in the journal Nature Medicine has looked at gut bacteria, immune responses and metabolism to find a way of diagnosing the condition.
The findings, potentially relevant to long Covid due to its similarity with chronic fatigue syndrome, come from data on 249 individuals analysed using a new artificial intelligence (AI) platform that identifies disease biomarkers from stool, blood, and other routine lab tests.
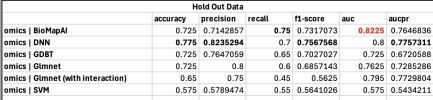
“Our study achieved 90 per cent accuracy in distinguishing individuals with chronic fatigue syndrome, which is significant because doctors currently lack reliable biomarkers for diagnosis,” said study author Dr Derya Unutmaz, professor in immunology at the Jackson Laboratory in the US.
“Some physicians doubt it as a real disease due to the absence of clear laboratory markers, sometimes attributing it to psychological factors.”
It is estimated that 404,000 people in the UK have chronic fatigue syndrome or ME, according to Action for ME. About half of the 1.9 million people in the UK with long Covid are also thought to have symptoms that are similar to ME.
Although it is not yet known what causes chronic fatigue syndrome, there is evidence that certain infections, including but not limited to viruses, can trigger the illness.
This new research, led by Dr Julia Oh, formerly at the Jackson Laboratory and now a microbiologist and professor at Duke University in North Carolina, investigates how the gut microbiome – the bacteria in your gut – and immune system interact in a patient with chronic fatigue syndrome.
To conduct the study, researchers used data collected from the Bateman Horne Center, a leading ME/CFS, long Covid and fibromyalgia research centre in Salt Lake City, Utah. Dr Ruoyun Xiong, also a lead author on the study, developed a research tool called BioMapAI.
annoying use of chronic fatigue as ever,This tool helped to compare gut bacteria, immune cells, blood test data, and clinical symptoms from 153 patients and 96 healthy participants over four years.
Researchers found analysing immune cells proved the most accurate in predicting how severe the participants’ symptoms were, but found data from gut bacteria helped predict participants’ emotional symptoms and sleep disturbances. They found those with chronic fatigue had lower levels of butyrate, a beneficial fatty acid produced in the gut, along with other nutrients essential for metabolism, inflammation control, and energy.