mariovitali
Senior Member (Voting Rights)
Dear All,
I am preparing an email to a number of researchers for the gene called ABCC6. Looking for further information regarding its function, one can find that it is a gene responsible for Pseudoxanthoma Elasticum (PXE) but the techniques i use suggest otherwise, meaning it should be further investigated.
Interestingly, a website discussing about PXE shows the following information in its FAQ :
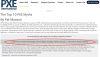
Please report on any of the SNPs shown below, despite the mention that an epidemiological study has been made between 1997 and 2001.
Here is the list of the SNPs. :
https://www.ncbi.nlm.nih.gov/snp/?term=rs2238472
Pathogenic
rs59513011 - Risk T
rs2606921 - Risk A
rs55778939 : Risk T
(New addition) rs58668703 - Risk T
(New addition) rs41278172 - Risk A
Likely benign :
rs72657698 : Risk C
Thank you for your help.
I am preparing an email to a number of researchers for the gene called ABCC6. Looking for further information regarding its function, one can find that it is a gene responsible for Pseudoxanthoma Elasticum (PXE) but the techniques i use suggest otherwise, meaning it should be further investigated.
Interestingly, a website discussing about PXE shows the following information in its FAQ :

Please report on any of the SNPs shown below, despite the mention that an epidemiological study has been made between 1997 and 2001.
Here is the list of the SNPs. :
https://www.ncbi.nlm.nih.gov/snp/?term=rs2238472
Pathogenic
rs59513011 - Risk T
rs2606921 - Risk A
rs55778939 : Risk T
(New addition) rs58668703 - Risk T
(New addition) rs41278172 - Risk A
Likely benign :
rs72657698 : Risk C
Thank you for your help.
Last edited:
