How Long COVID develops is still largely unknown. New molecular connections are revealed in a recent study led by the Centre for Individualised Infection Medicine (CiiM). Using their approach of examining different molecular levels within individual cells, the researchers identified a specific...
www.eurekalert.org
News Release 29-Jan-2026
How does Long COVID develop? New piece of the puzzle found
Big data study finds link to molecular cell state of immune cells
Peer-Reviewed Publication
Helmholtz Centre for Infection Research
FacebookXLinkedInWeChatBlueskyMessageWhatsAppEmail
After infection with the SARS-CoV-2 virus, up to ten percent of those affected in Germany develop Long COVID. The symptoms associated with it, such as fatigue, concentration problems, respiratory issues, or neurological problems, can last for months or years. Furthermore, the clinical picture can differ from person to person. “Long COVID is an extremely complex disease with various manifestations,” says Prof. Yang Li, head of the “Computational Biology for Individualised Medicine” department and director of CiiM. “How and to what extent Long COVID develops is still largely unknown. Figuratively speaking, we are unfortunately only looking at an extremely incomplete puzzle.”
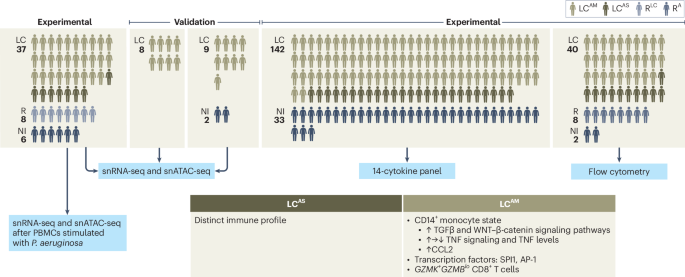
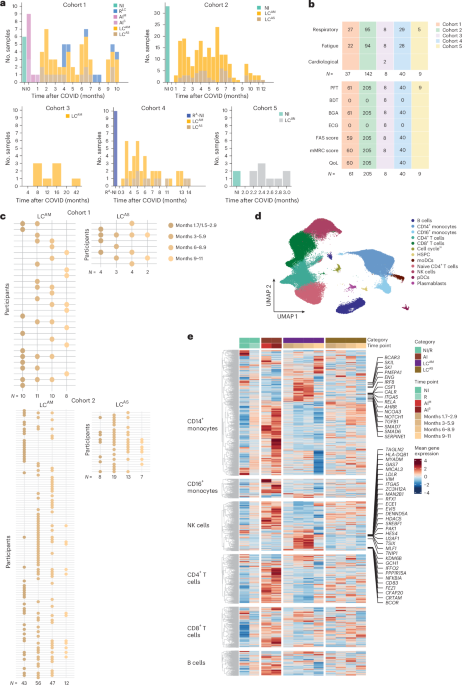
The research team led by study director Yang Li, together with the teams of Prof. Thomas Illig (MHH) and Prof. Jie Sun (University of Virginia, USA), as well as other cooperation partners, set out to find further pieces of the puzzle that could help uncover the disease-causing mechanisms behind Long COVID. To this end, the researchers took a closer look at immune cells from patients with Long COVID in their study. The samples came from MHH's central biobank. "We examined the cells using a so-called single-cell multiomics approach. This allowed us to record the molecules’ status within a cell and gain insights into its cellular relationships," explains Dr Saumya Kumar, CiiM scientist and first author of the study. The researchers also determined the content of pro-inflammatory messenger substances, known as cytokines, in blood plasma. “The central and innovative approach of our study is to classify patient data according to the severity of the original COVID-19 disease,” says Yang Li. “This approach allowed us to capture the associated molecular differences in immune response across patients. Only in this way, clear molecular characteristics underlying the chronic symptoms of Long COVID could be identified.”
How does the molecular setting in immune cells change over the course of Long COVID? Are there clear molecular markers associated with the severity of fatigue or respiratory symptoms? Researchers investigated these and other questions in their big data study. And what then came into focus for the researchers from this extensive treasure trove of data was a specific molecular state of so-called CD14+ monocytes. These immune cells belong to the white blood cells and are an important part of the immune defense. “With the help of single-cell analysis, we were able to zoom in on these cells. This revealed that monocytes with a specific molecular state (i.e. molecular profile), which we called “LC-Mo”, were particularly prevalent in Long COVID patients who had previously experienced mild to moderate COVID-19 disease,” says Saumya Kumar. “In addition, LC-Mo correlated with the severity of fatigue and respiratory symptoms and was associated with elevated cytokine levels in blood plasma, which are an indicator of inflammatory processes in the body.”
With LC-Mo, the researchers have thus found an important new piece of the puzzle. “Its exact place in the pathogenesis of Long COVID has yet to be determined, but it offers exciting starting points for further studies, for example, with regard to genetic risk factors or individualised medicine,” says Yang Li. "If we can gain a better understanding of the background to the development of Long COVID, it will also help us to better understand the development of possible late or long-term consequences of other infectious diseases."
The research was funded by an ERC Starting Grant (ModVaccine), the COVID-19 Research Network of Lower Saxony (COFONI) and the Lower Saxony Centre for AI & Causal Methods in Medicine (CAIMed), both with funds from the Lower Saxony Ministry of Science and Culture (MWK), as well as the Federal Ministry of Research, Technology and Space (BMFTR).
Text: Nicole Silbermann
Centre for Individualised Infection Medicine:
The CiiM is a joint venture of the Helmholtz Centre for Infection Research (HZI) and Hannover Medical School (MHH) and is the first research institute explicitly dedicated to develop personalized medicine for infections. The departments and groups of the CiiM are working towards the vision of treating infectious patients in an adapted and optimized way according to the specific needs and requirements of each individual. To achieve this, CiiM is dedicated to researching individual characteristics and their impact on susceptibility to infection or treatment success with available therapies.
https://www.ciim-hannover.de/en/
Helmholtz Centre for Infection Research:
Scientists at the Helmholtz Centre for Infection Research (HZI) in Braunschweig and its other sites in Germany are engaged in the study of bacterial and viral infections and the body’s defense mechanisms. They have a profound expertise in natural compound research and its exploitation as a valuable source for novel anti-infectives. As member of the Helmholtz Association and the German Center for Infection Research (DZIF) the HZI performs translational research laying the ground for the development of new treatments and vaccines against infectious diseases.
https://www.helmholtz-hzi.de/en
Journal
Nature Immunology
DOI
10.1038/s41590-025-02387-1
Method of Research
Data/statistical analysis
Subject of Research
Human tissue samples
Article Title
A distinct monocyte transcriptional state links systemic immune dysregulation to pulmonary impairment in long COVID
Article Publication Date
14-Jan-2026