jnmaciuch
Senior Member (Voting Rights)
Mitochondria-localised ZNFX1 functions as a dsRNA sensor to initiate antiviral responses through MAVS
Abstract
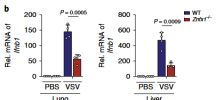
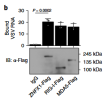
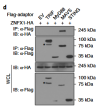
In the past two decades, emerging studies have suggested that DExD/H box helicases belonging to helicase superfamily 2 (SF2) play essential roles in antiviral innate immunity. However, the antiviral functions of helicase SF1, which shares a conserved helicase core with SF2, are little understood. Here we demonstrate that zinc finger NFX1-type containing 1 (ZNFX1), a helicase SF1, is an interferon (IFN)-stimulated, mitochondrial-localised dsRNA sensor that specifically restricts the replication of RNA viruses. Upon virus infection, ZNFX1 immediately recognizes viral RNA through its Armadillo-type fold and P-loop domain and then interacts with mitochondrial antiviral signalling protein to initiate the type I IFN response without depending on retinoic acid-inducible gene I-like receptors (RLRs). In short, as is the case with interferon-stimulated genes (ISGs) alone, ZNFX1 can induce IFN and ISG expression at an early stage of RNA virus infection to form a positively regulated loop of the well-known RLR signalling. This provides another layer of understanding of the complexity of antiviral immunity.
Web | DOI | PDF | Nature Cell Biology
Wang, Yao; Yuan, Shaochun; Jia, Xin; Ge, Yong; Ling, Tao; Nie, Meng; Lan, Xihong; Chen, Shangwu; Xu, Anlong
Abstract
In the past two decades, emerging studies have suggested that DExD/H box helicases belonging to helicase superfamily 2 (SF2) play essential roles in antiviral innate immunity. However, the antiviral functions of helicase SF1, which shares a conserved helicase core with SF2, are little understood. Here we demonstrate that zinc finger NFX1-type containing 1 (ZNFX1), a helicase SF1, is an interferon (IFN)-stimulated, mitochondrial-localised dsRNA sensor that specifically restricts the replication of RNA viruses. Upon virus infection, ZNFX1 immediately recognizes viral RNA through its Armadillo-type fold and P-loop domain and then interacts with mitochondrial antiviral signalling protein to initiate the type I IFN response without depending on retinoic acid-inducible gene I-like receptors (RLRs). In short, as is the case with interferon-stimulated genes (ISGs) alone, ZNFX1 can induce IFN and ISG expression at an early stage of RNA virus infection to form a positively regulated loop of the well-known RLR signalling. This provides another layer of understanding of the complexity of antiviral immunity.
Web | DOI | PDF | Nature Cell Biology