Identification of an immunological signature of long COVID syndrome
Guerrera, Gisella; Sambucci, Manolo; Timperi, Eleonora; Picozza, Mario; Misiti, Andrea; Placido, Roberta; Corbisiero, Silvia; D’Orso, Silvia; Termine, Andrea; Fabrizio, Carlo; Gargano, Francesca; Eleuteri, Sharon; Marchioni, Luisa; Bordoni, Veronica; Coppola, Luigi; Iannetta, Marco; Agrati, Chiara; Borsellino, Giovanna; Battistini, Luca
INTRODUCTION
Acute COVID-19 infection causes significant alterations in the innate and adaptive immune systems. While most individuals recover naturally, some develop long COVID (LC) syndrome, marked by persistent or new symptoms weeks to months after SARS-CoV-2 infection. Despite its prevalence, there are no clinical tests to distinguish LC patients from those fully recovered. Understanding the immunological basis of LC is essential for improving diagnostic and treatment approaches.
METHODS
We performed deep immunophenotyping and functional assays to examine the immunological profiles of LC patients, individuals with active COVID-19, recovered patients, and healthy donors. This analysis assessed both innate and adaptive immune features, identifying potential biomarkers for LC syndrome. A Binomial Generalized Linear Model (BGLM) was used to pinpoint immune features characterizing LC.
RESULTS
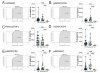
COVID-19 patients exhibited depletion of innate immune cell subsets, including plasmacytoid and conventional dendritic cells, classical, non-classical, and intermediate monocytes, and monocyte-derived inflammatory dendritic cells. Elevated basal inflammation was observed in COVID-19 patients compared to LC patients, whose immune profiles were closer to those of healthy donors and recovered individuals. However, LC patients displayed persistent immune alterations, including reduced T cell subsets (CD4, CD8, Tregs) and switched memory B cells, similar to COVID-19 patients. Through BGLM, a unique adaptive immune signature for LC was identified, featuring memory CD8 and gd T cells with low proliferative capacity and diminished expression of activation and homing receptors.
DISCUSSION
The findings highlight a unique immunological signature associated with LC syndrome, characterized by persistent adaptive immune dysregulation. While LC patients displayed recovery in innate immune profiles comparable to healthy and Recovered individuals, deficits in T cell and memory B cell populations were evident, differentiating LC from full recovery. These findings provide insights into LC pathogenesis and may support the development of diagnostic tools and targeted therapies.
Link | PDF (Frontiers in Immunology) [Open Access]
Guerrera, Gisella; Sambucci, Manolo; Timperi, Eleonora; Picozza, Mario; Misiti, Andrea; Placido, Roberta; Corbisiero, Silvia; D’Orso, Silvia; Termine, Andrea; Fabrizio, Carlo; Gargano, Francesca; Eleuteri, Sharon; Marchioni, Luisa; Bordoni, Veronica; Coppola, Luigi; Iannetta, Marco; Agrati, Chiara; Borsellino, Giovanna; Battistini, Luca
INTRODUCTION
Acute COVID-19 infection causes significant alterations in the innate and adaptive immune systems. While most individuals recover naturally, some develop long COVID (LC) syndrome, marked by persistent or new symptoms weeks to months after SARS-CoV-2 infection. Despite its prevalence, there are no clinical tests to distinguish LC patients from those fully recovered. Understanding the immunological basis of LC is essential for improving diagnostic and treatment approaches.
METHODS
We performed deep immunophenotyping and functional assays to examine the immunological profiles of LC patients, individuals with active COVID-19, recovered patients, and healthy donors. This analysis assessed both innate and adaptive immune features, identifying potential biomarkers for LC syndrome. A Binomial Generalized Linear Model (BGLM) was used to pinpoint immune features characterizing LC.
RESULTS
COVID-19 patients exhibited depletion of innate immune cell subsets, including plasmacytoid and conventional dendritic cells, classical, non-classical, and intermediate monocytes, and monocyte-derived inflammatory dendritic cells. Elevated basal inflammation was observed in COVID-19 patients compared to LC patients, whose immune profiles were closer to those of healthy donors and recovered individuals. However, LC patients displayed persistent immune alterations, including reduced T cell subsets (CD4, CD8, Tregs) and switched memory B cells, similar to COVID-19 patients. Through BGLM, a unique adaptive immune signature for LC was identified, featuring memory CD8 and gd T cells with low proliferative capacity and diminished expression of activation and homing receptors.
DISCUSSION
The findings highlight a unique immunological signature associated with LC syndrome, characterized by persistent adaptive immune dysregulation. While LC patients displayed recovery in innate immune profiles comparable to healthy and Recovered individuals, deficits in T cell and memory B cell populations were evident, differentiating LC from full recovery. These findings provide insights into LC pathogenesis and may support the development of diagnostic tools and targeted therapies.
Link | PDF (Frontiers in Immunology) [Open Access]