Chandelier
Senior Member (Voting Rights)
Development of an ultrahigh affinity, trimeric ACE2 biologic as a universal SARS-CoV-2 antagonist - Communications Biology
Structure-guided engineering of a trimeric ACE2 viral decoy reveals high affinity for spike protein, viral inhibition, and serum stability, offering a universal therapeutic strategy against SARS-CoV-2 variants and SARS-CoV-1 through symmetry-matched avidity.
Juliet Gonzales, Tynan Young, Hyeran Choi, Miso Park, Yead Jewel, Chengcheng Fan, Rahul Purohit, Pamela J. Bjorkman & John C. Williams
Abstract
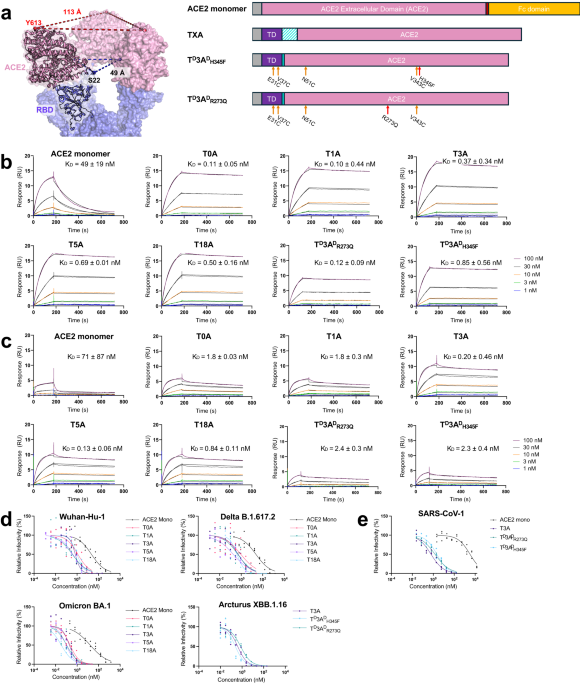
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for the COVID-19 pandemic, utilizes membrane-bound, angiotensin-converting enzyme II (ACE2) for internalization and infection.We describe the development of a biologic that takes advantage of the proximity of the N-terminus of bound ACE2 to the three-fold symmetry axis of the spike protein to create an ultrapotent, trivalent ACE2 entry antagonist.
Distinct disulfide bonds were added to enhance serum stability and a single point mutation was introduced to eliminate enzymatic activity.
Through surface plasmon resonance, pseudovirus neutralization assays, and single-particle cryo-electron microscopy, we show this antagonist binds to and inhibits SARS-CoV-2 variants.
We further show the antagonist binds to and inhibits a 2003 SARS-CoV-1 strain.
Collectively, structural insight has allowed us to design a universal trivalent antagonist against all variants of SARS-CoV-2 tested, suggesting it will be active against the emergence of future mutants.
